4ZV2
 
 | |
1A28
 
 | |
6XR4
 
 | |
1MR0
 
 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | Deposit date: | 2002-09-17 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
6XGU
 
 | | Crystal Structure of KRAS-Q61R (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF | | Descriptor: | GLYCEROL, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6RAK
 
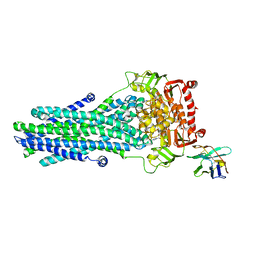 | | Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
5W3J
 
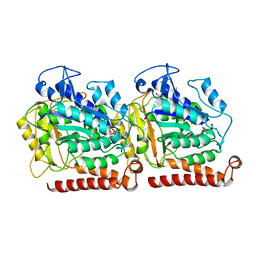 | | Yeast microtubule stabilized with Taxol assembled from mutated tubulin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
8CQB
 
 | |
4ZSV
 
 | |
1MTN
 
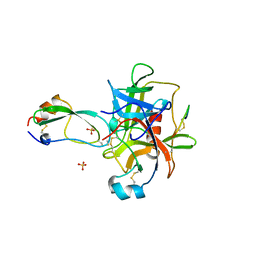 | | BOVINE ALPHA-CHYMOTRYPSIN:BPTI CRYSTALLIZATION | | Descriptor: | ALPHA-CHYMOTRYPSIN, BASIC PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Capasso, C, Rizzi, M, Menegatti, E, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-08-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the bovine alpha-chymotrypsin:Kunitz inhibitor complex. An example of multiple protein:protein recognition sites.
J.Mol.Recog., 10, 1997
|
|
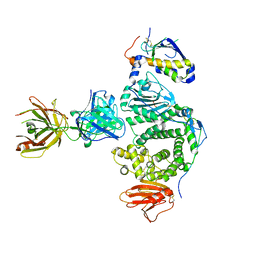
5W0K
 
 | |
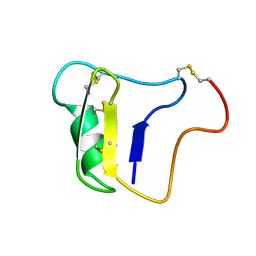
1VNA
 
 | |
4ZSZ
 
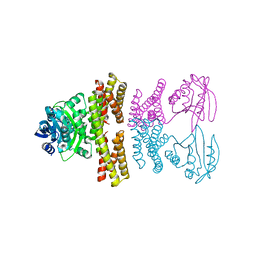 | |
5W1F
 
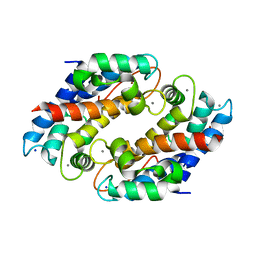 | | Crystal structure of Ni(II)- and Ca(II)-bound human calprotectin | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Protein S100-A8, ... | | Authors: | Nakashige, T.G, Drennan, C.L, Nolan, E.M. | | Deposit date: | 2017-06-03 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nickel Sequestration by the Host-Defense Protein Human Calprotectin.
J. Am. Chem. Soc., 139, 2017
|
|
1VNB
 
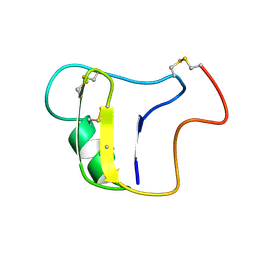 | |
6XBZ
 
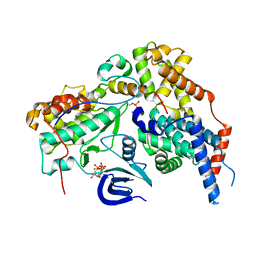 | | Structure of the human CDK-activating kinase | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Greber, B.J, Perez-Bertoldi, J.M, Lim, K, Iavarone, A.T, Toso, D.B, Nogales, E. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The cryoelectron microscopy structure of the human CDK-activating kinase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3VBT
 
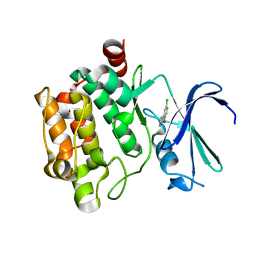 | |
4ZTE
 
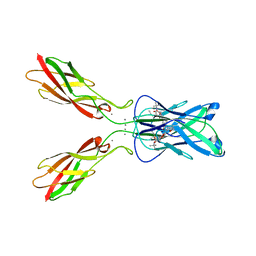 | | Crystal structure of human E-Cadherin (residues 3-213) in complex with a peptidomimetic inhibitor | | Descriptor: | CALCIUM ION, Cadherin-1, N-{[(2S,5S)-1-benzyl-5-(2-{[(2S,3S)-1-(tert-butylamino)-3-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl)-3,6-dioxopiperazin-2-yl]methyl}-L-alpha-asparagine | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
5W5B
 
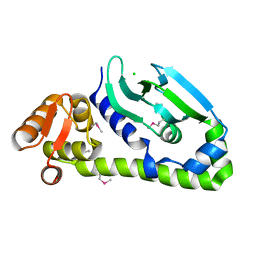 | | Crystal structure of Mycobacterium tuberculosis CRP-FNR family transcription factor Cmr (Rv1675c), truncated construct | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator Cmr | | Authors: | Cheung, J, Cassidy, M, Ginter, C, Ranganathan, S, Pata, D.J, McDonough, K.A. | | Deposit date: | 2017-06-14 | | Release date: | 2017-12-13 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural features drive DNA binding properties of Cmr, a CRP family protein in TB complex mycobacteria.
Nucleic Acids Res., 46, 2018
|
|
5W2A
 
 | |
4V8Q
 
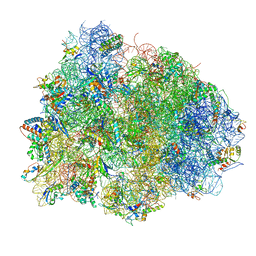 | | Complex of SmpB, a tmRNA fragment and EF-Tu-GDP-Kirromycin with the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Neubauer, C, Gillet, R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2011-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Decoding in the absence of a codon by tmRNA and SmpB in the ribosome.
Science, 335, 2012
|
|
5W2J
 
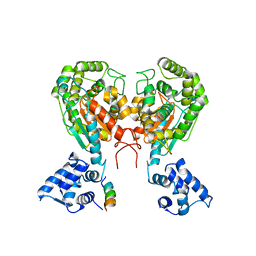 | | Crystal structure of dimeric form of mouse Glutaminase C | | Descriptor: | CHLORIDE ION, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Cerione, R.A, Li, Y. | | Deposit date: | 2017-06-06 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic Basis of Glutaminase Activation: A KEY ENZYME THAT PROMOTES GLUTAMINE METABOLISM IN CANCER CELLS.
J. Biol. Chem., 291, 2016
|
|
4V83
 
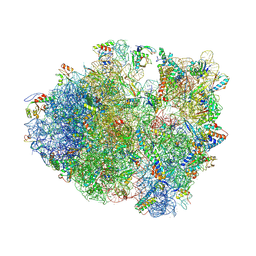 | | Crystal structure of a complex containing domain 3 from the PSIV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6UX2
 
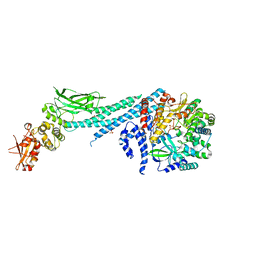 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5W3O
 
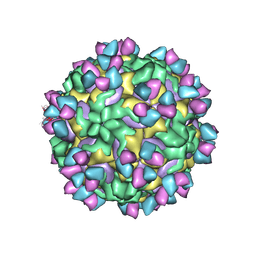 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, empty particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
