5B2G
 
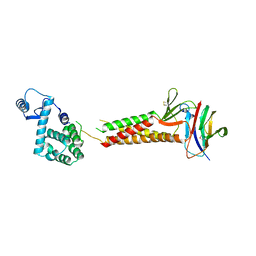 | | Crystal structure of human claudin-4 in complex with C-terminal fragment of Clostridium perfringens enterotoxin | | Descriptor: | Endolysin,Claudin-4, Heat-labile enterotoxin B chain | | Authors: | Shinoda, T, Kimura-Someya, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin
Sci Rep, 6, 2016
|
|
6GIQ
 
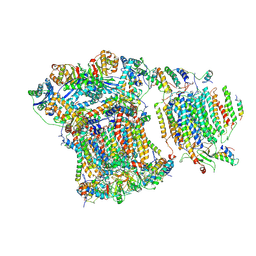 | | Saccharomyces cerevisiae respiratory supercomplex III2IV | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, ... | | Authors: | Rathore, S, Berndtsson, J, Conrad, J, Ott, M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-01-02 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of the yeast respiratory supercomplex.
Nat. Struct. Mol. Biol., 26, 2019
|
|
5B4Y
 
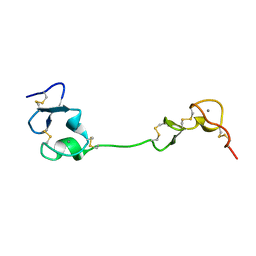 | | Crystal structure of the LA12 fragment of ApoER2 | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 8 | | Authors: | Nogi, T, Tabata, S, Hirai, H, Yasui, N, Takagi, J. | | Deposit date: | 2016-04-20 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ectodomain from a LDLR close homologue in complex with its physiological ligand.
To Be Published
|
|
5B5G
 
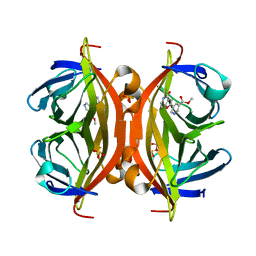 | | Crystal structure of ALiS4-Streptavidin complex | | Descriptor: | SULFITE ION, Streptavidin, methyl 5-(4-oxidanylidene-5~{H}-furo[3,2-c]pyridin-2-yl)pyridine-3-carboxylate | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
6GL8
 
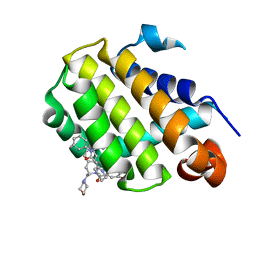 | | Crystal structure of Bcl-2 in complex with the novel orally active inhibitor S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Casara, P, Davidson, J, Claperon, A, Le Toumelin-Braizat, G, Vogler, M, Bruno, A, Chanrion, M, Lysiak-Auvity, G, Le Diguarher, T, Starck, J.B, Chen, I, Whitehead, N, Graham, C, Matassova, N, Dokurno, P, Pedder, C, Wang, Y, Qiu, S, Girard, A.M, Schneider, E, Grave, F, Studeny, A, Guasconi, G, Rocchetti, F, Maiga, S, Henlin, J.M, Colland, F, Kraus-Berthier, L, Le Gouill, S, Dyer, M.J.S, Hubbard, R, Wood, M, Amiot, M, Cohen, G.M, Hickman, J.A, Morris, E, Murray, J, Geneste, O. | | Deposit date: | 2018-05-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S55746 is a novel orally active BCL-2 selective and potent inhibitor that impairs hematological tumor growth.
Oncotarget, 9, 2018
|
|
5AW7
 
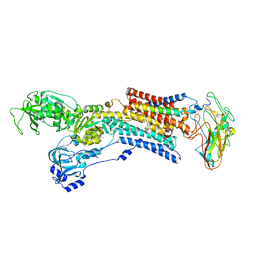 | | Kinetics by X-ray crystallography: Rb+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 11.3 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
6GML
 
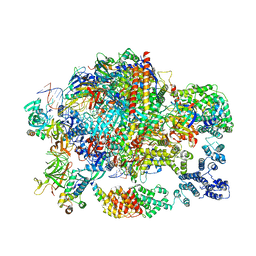 | | Structure of paused transcription complex Pol II-DSIF-NELF | | Descriptor: | DNA-directed RNA polymerase II subunit RPB9, DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Vos, S.M, Farnung, L, Urlaub, H, Cramer, P. | | Deposit date: | 2018-05-27 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of paused transcription complex Pol II-DSIF-NELF.
Nature, 560, 2018
|
|
5AVS
 
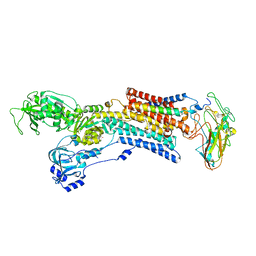 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 3.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW4
 
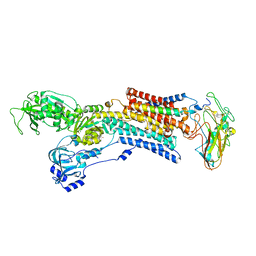 | | Kinetics by X-ray crystallography: Rb+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 1.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AYM
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in outward-facing state with soaked iron | | Descriptor: | FE (II) ION, Solute carrier family 39 (Iron-regulated transporter) | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
5AX8
 
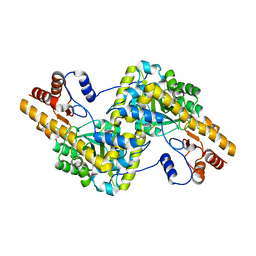 | | Recombinant expression, purification and preliminary crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, mitochondrial | | Authors: | Jiang, X, Wang, J, Chang, H, Zhou, Y. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Recombinant expression, purification and crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase
Biosci Trends, 10, 2016
|
|
5AZB
 
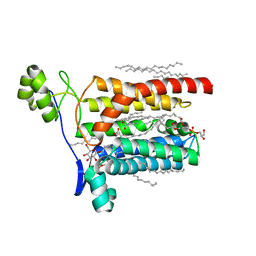 | | Crystal structure of Escherichia coli Lgt in complex with phosphatidylglycerol and the inhibitor palmitic acid | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PALMITIC ACID, Prolipoprotein diacylglyceryl transferase, ... | | Authors: | Zhang, X.C, Mao, G, Zhao, Y. | | Deposit date: | 2015-09-30 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of E. coli lipoprotein diacylglyceryl transferase
Nat Commun, 7, 2016
|
|
6GWF
 
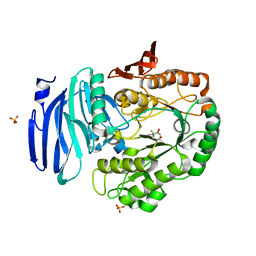 | | Alpha-galactosidase mutant D387A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 2,4-dinitro leaving group | | Descriptor: | (1~{S},2~{S},5~{S},6~{R})-5-(2,4-dinitrophenoxy)-6-fluoranyl-3-(hydroxymethyl)cyclohex-3-ene-1,2-diol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
5B3S
 
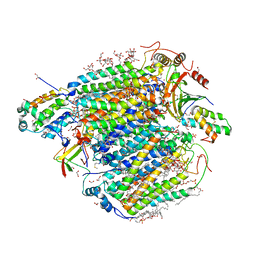 | | Bovine heart cytochrome c oxidase in the carbon monoxide-bound mixed-valence state at 1.68 angstrom resolution (50 K) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Shinzawa-Ito, K, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2016-03-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Bovine heart cytochrome c oxidase in the carbon monoxide-bound mixed-valence state at 1.68 angstrom resolution (50 K)
To Be Published
|
|
5B35
 
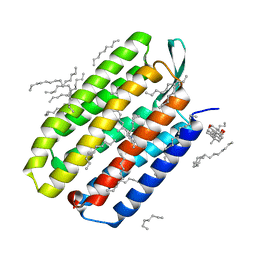 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T, Suzuki, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B5F
 
 | | Crystal structure of ALiS3-Streptavidin complex | | Descriptor: | N-methyl-3-(4-oxo-4,5-dihydrofuro[3,2-c]pyridin-2-yl)benzenesulfonamide, Streptavidin | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
6FRY
 
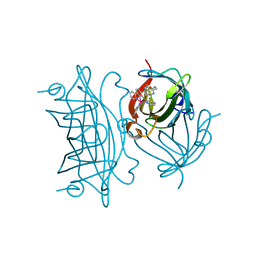 | | Photo-Driven Hydrogen Evolution by an Artificial Hydrogenase Utilizing the Biotin-Streptavidin Technology | | Descriptor: | Streptavidin, [CoBr(appy)-Biot]Br | | Authors: | Keller, S, Probst, B, Heinisch, T, Alberto, R, Ward, T.R. | | Deposit date: | 2018-02-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Photo-Driven Hydrogen Evolution by an Artificial Hydrogenase Utilizing the Biotin-Streptavidin Technology
Helv.Chim.Acta, 101, 2018
|
|
6G2J
 
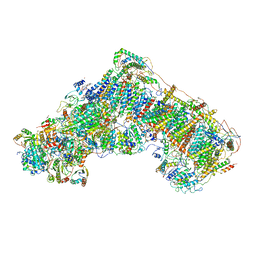 | | Mouse mitochondrial complex I in the active state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Agip, A.N.A, Blaza, J.N, Bridges, H.R, Viscomi, C, Rawson, S, Muench, S.P, Hirst, J. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-06 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6FO6
 
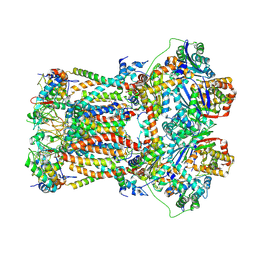 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial inhibitor SCR0911 | | Descriptor: | 7-methoxy-3-methyl-2-[5-[4-(trifluoromethyloxy)phenyl]pyridin-3-yl]quinolin-4-ol, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6G7L
 
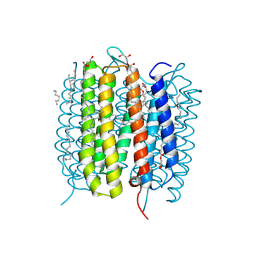 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 8.3 ms state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G0V
 
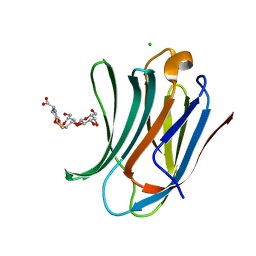 | | Human Galectin-3 in complex with a TF tumor-associated antigen mimetic | | Descriptor: | (3~{R},5~{R},6~{S},7~{S},8~{R},13~{S})-5-(hydroxymethyl)-7-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-oxidanyl-11-oxidanylidene-2,4-dioxa-9-thia-12-azatricyclo[8.4.0.0^{3,8}]tetradec-1(10)-ene-13-carboxylic acid, CHLORIDE ION, Galectin-3 | | Authors: | Trovao, F.G, Santarsia, S, Carvalho, A.L. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular Recognition of a Thomsen-Friedenreich Antigen Mimetic Targeting Human Galectin-3.
ChemMedChem, 13, 2018
|
|
5DFF
 
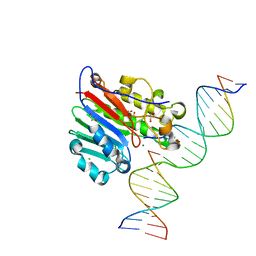 | | Human APE1 product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5CXV
 
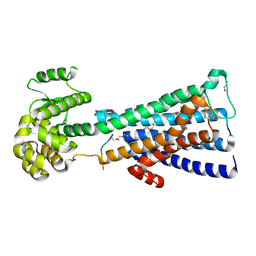 | | Structure of the human M1 muscarinic acetylcholine receptor bound to antagonist Tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, 1,2-ETHANEDIOL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, B, Feng, D, Li, X, Kobilka, T.S, Kobilka, B.K. | | Deposit date: | 2015-07-29 | | Release date: | 2016-03-09 | | Last modified: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the M1 and M4 muscarinic acetylcholine receptors.
Nature, 531, 2016
|
|
6GA3
 
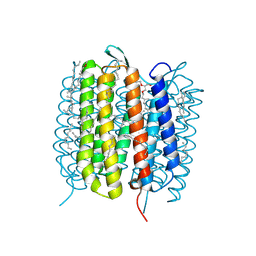 | | Bacteriorhodopsin, 33 ms state, ensemble refinement | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAC
 
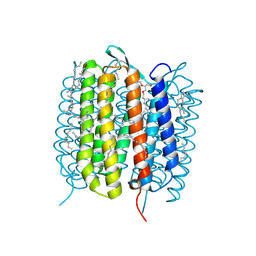 | | BACTERIORHODOPSIN, 490 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
