7XA4
 
 | |
7XA2
 
 | | Thermotoga maritima ferritin variant-Tm-E(S111H) | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Liu, Y, Zhao, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Directed Self-Assembly of Dimeric Building Blocks into Networklike Protein Origami to Construct Hydrogels.
Acs Nano, 16, 2022
|
|
5GT8
 
 | | Crystal Structure of apo-CASTOR1 | | Descriptor: | GATS-like protein 3 | | Authors: | Guo, L, Deng, D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states
Biochem. Biophys. Res. Commun., 508, 2019
|
|
4R7W
 
 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, 1,2-ETHANEDIOL, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine
To be Published
|
|
5WO4
 
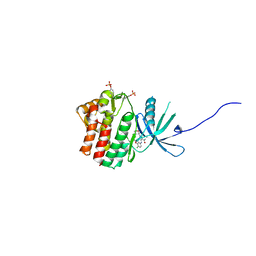 | | JAK1 complexed with compound 28 | | Descriptor: | 3-[(4-chloro-3-methoxyphenyl)amino]-1-[(3R,4S)-4-cyanooxan-3-yl]-1H-pyrazole-4-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lesburg, C.A, Patel, S.B. | | Deposit date: | 2017-08-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Discovery of 3-((4-Chloro-3-methoxyphenyl)amino)-1-((3R,4S)-4-cyanotetrahydro-2H-pyran-3-yl)-1H-pyrazole-4-carboxamide, a Highly Ligand Efficient and Efficacious Janus Kinase 1 Selective Inhibitor with Favorable Pharmacokinetic Properties.
J. Med. Chem., 60, 2017
|
|
4WSA
 
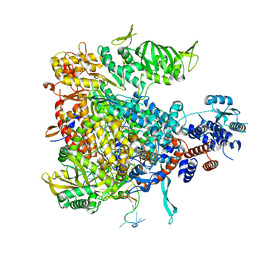 | | Crystal structure of Influenza B polymerase bound to the vRNA promoter (FluB1 form) | | Descriptor: | Influenza B vRNA promoter 3' end, Influenza B vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
4WWZ
 
 | | UndA complexed with 2,3-dodecenoic acid | | Descriptor: | (2E)-dodec-2-enoic acid, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | Li, X, Cate, J.D.H. | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Microbial biosynthesis of medium-chain 1-alkenes by a nonheme iron oxidase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WSB
 
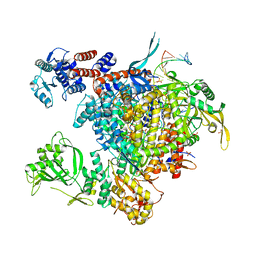 | | Bat Influenza A polymerase with bound vRNA promoter | | Descriptor: | Influenza A polymerase vRNA promoter 3' end, Influenza A polymerase vRNA promoter 5' end, PHOSPHATE ION, ... | | Authors: | Cusack, S, Pflug, A, Guilligay, D, Reich, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
5FDA
 
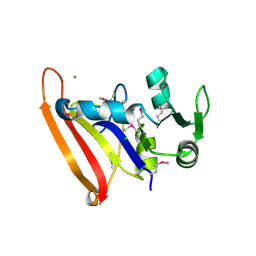 | | The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | structure of dihydrofolate reductase from Yersinia pestis complex with
To Be Published
|
|
7LBC
 
 | | Structure of human GGT1 in complex with Lnt2-65 compound | | Descriptor: | (2R)-2-[(2-aminoethyl)amino]-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-07 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
5YXB
 
 | | A ligand binding to FXR | | Descriptor: | 2-methoxyethyl (2E)-3-phenylprop-2-en-1-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A ligand binding to FXR
To Be Published
|
|
5YXL
 
 | | A ligand M binding to FXR | | Descriptor: | 2-methoxyethyl propan-2-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A ligand M binding to FXR
To Be Published
|
|
6FUC
 
 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 | | Descriptor: | Aminoglycoside phosphotransferase | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
4L4Z
 
 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | Descriptor: | (2S)-2,3,3-trihydroxy-4-oxopentyl dihydrogen phosphate, Transcriptional regulator LsrR | | Authors: | Ryu, K.S, Ha, J.H, Eo, Y. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
6FUX
 
 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 in complex with ADP and streptomycin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, GLYCEROL, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6MYV
 
 | | Sialidase26 co-crystallized with DANA-Gc | | Descriptor: | 2,6-anhydro-3,5-dideoxy-5-[(hydroxyacetyl)amino]-D-glycero-L-altro-non-2-enonic acid, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Huang, J, Siegel, D, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
1B3O
 
 | | TERNARY COMPLEX OF HUMAN TYPE-II INOSINE MONOPHOSPHATE DEHYDROGENASE WITH 6-CL-IMP AND SELENAZOLE ADENINE DINUCLEOTIDE | | Descriptor: | 6-CHLOROPURINE RIBOSIDE, 5'-MONOPHOSPHATE, PROTEIN (INOSINE MONOPHOSPHATE DEHYDROGENASE 2), ... | | Authors: | Colby, T.D, Vanderveen, K, Strickler, M.D, Goldstein, B.M. | | Deposit date: | 1998-12-14 | | Release date: | 1999-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human type II inosine monophosphate dehydrogenase: implications for ligand binding and drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4L50
 
 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | Descriptor: | (2S)-2,3,3-trihydroxy-6-methyl-4-oxoheptyl dihydrogen phosphate, Transcriptional regulator LsrR | | Authors: | Ryu, K.S, Ha, J.H, Eo, Y. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
4CQ5
 
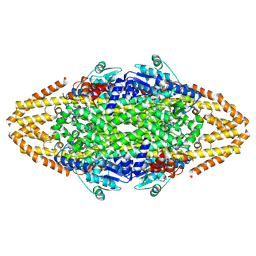 | | Structural Investigations into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus chinensis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHENYLALANINE AMINOMUTASE, ... | | Authors: | Wybenga, G.G, Szymanski, W, Wu, B, Feringa, B.L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2014-02-11 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Investigations Into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus Chinensis.
Biochemistry, 53, 2014
|
|
4CMM
 
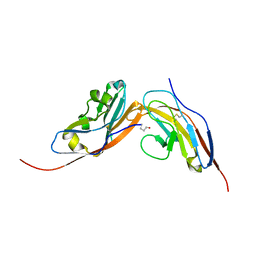 | | Structure of human CD47 in complex with human Signal Regulatory Protein (SIRP) alpha v1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUKOCYTE SURFACE ANTIGEN CD47, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE SUBSTRATE 1 | | Authors: | Hatherley, D, Lea, S.M, Johnson, S, Barclay, A.N. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Polymorphisms in the Human Inhibitory Signal-Regulatory Protein Alpha Do not Affect Binding to its Ligand Cd47.
J.Biol.Chem., 289, 2014
|
|
7LD9
 
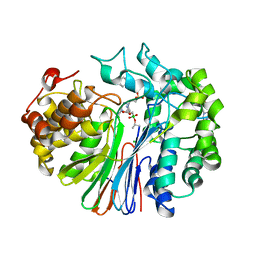 | | Structure of human GGT1 in complex with ABBA | | Descriptor: | (2R)-2-amino-4-boronobutanoic acid, (2S)-2-amino-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-12 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
1AN9
 
 | | D-AMINO ACID OXIDASE COMPLEX WITH O-AMINOBENZOATE | | Descriptor: | 2-AMINOBENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Miura, R, Setoyama, C, Nishina, Y, Shiga, K, Mizutani, H, Miyahara, I, Hirotsu, K. | | Deposit date: | 1997-06-28 | | Release date: | 1997-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic studies on D-amino acid oxidase x substrate complex: implications of the crystal structure of enzyme x substrate analog complex.
J.Biochem.(Tokyo), 122, 1997
|
|
5GT7
 
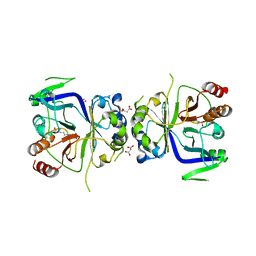 | | Crystal Structure of Arg-bound CASTOR1 | | Descriptor: | ARGININE, GATS-like protein 3, MALONATE ION | | Authors: | Guo, L, Deng, D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states
Biochem. Biophys. Res. Commun., 508, 2019
|
|
4J3L
 
 | | Tankyrase 2 in complex with 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide | | Descriptor: | 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
5AUM
 
 | |
