7YBX
 
 | | Crystal structure of FGFR4(V550M) kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
5K9Y
 
 | | Crystal structure of a thermophilic xylanase A from Bacillus subtilis 1A1 quadruple mutant Q7H/G13R/S22P/S179C | | Descriptor: | Endo-1,4-beta-xylanase A | | Authors: | Pinheiro, M.P, Ferreira, T.L, Silva, S.R.B, Fuzo, C.A, Silva, S.R, Lourenzoni, M.R, Vieira, D.S, Ward, R.J, Nonato, M.C. | | Deposit date: | 2016-06-01 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The role of local residue environmental changes in thermostable mutants of the GH11 xylanase from Bacillus subtilis.
Int. J. Biol. Macromol., 97, 2017
|
|
5K9B
 
 | | Azotobacter vinelandii Flavodoxin II | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin-2, MAGNESIUM ION | | Authors: | Rees, D.C, Segal, H.M, Spatzal, T. | | Deposit date: | 2016-05-31 | | Release date: | 2017-08-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.174 Å) | | Cite: | Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II.
Protein Sci., 26, 2017
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
7YKE
 
 | | Crystal structure of chondroitin ABC lyase I in complex with chondroitin disaccharide 4,6-sulfate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4,6-di-O-sulfo-beta-D-galactopyranose, Chondroitin sulfate ABC endolyase, MAGNESIUM ION | | Authors: | Takashima, M, Watanabe, I, Miyanaga, A, Eguchi, T. | | Deposit date: | 2022-07-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Biochemical and crystallographic assessments of the effect of 4,6-O-disulfated disaccharide moieties in chondroitin sulfate E on chondroitinase ABC I activity.
Febs J., 290, 2023
|
|
7YI7
 
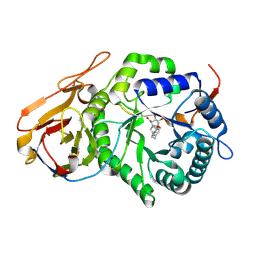 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-[4-(trifluoromethyl)phenyl]ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lead identification of novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid derivative as a potent heparanase-1 inhibitor.
Bioorg.Med.Chem.Lett., 79, 2022
|
|
5KHP
 
 | | Tightening the Recognition of Tetravalent Zr and Th Complexes by the Siderophore-Binding Mammalian Protein Siderocalin for Theranostic Applications | | Descriptor: | GLYCEROL, N,N'-(butane-1,4-diyl)bis(N-{3-[(2,3-dihydroxybenzene-1-carbonyl)amino]propyl}-2,3-dihydroxybenzamide), Neutrophil gelatinase-associated lipocalin, ... | | Authors: | Rupert, P.B, Strong, R.K. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered Recognition of Tetravalent Zirconium and Thorium by Chelator-Protein Systems: Toward Flexible Radiotherapy and Imaging Platforms.
Inorg Chem, 55, 2016
|
|
7YJC
 
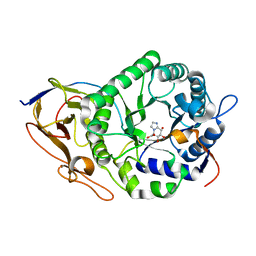 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lead identification of novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid derivative as a potent heparanase-1 inhibitor.
Bioorg.Med.Chem.Lett., 79, 2022
|
|
5KEZ
 
 | |
5KGR
 
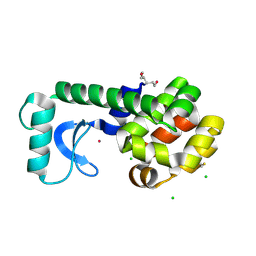 | | Spin-Labeled T4 Lysozyme Construct I9V1/V131V1 (30 days) | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-06-13 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
7XVH
 
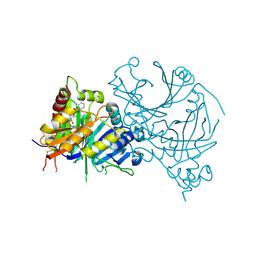 | | Crystal structure of AtHPPD-Y13287 complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-05-23 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
5KIV
 
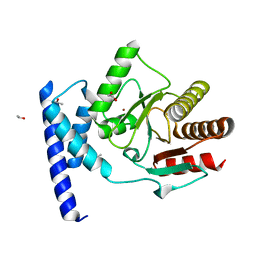 | | Crystal structure of SauMacro (SAV0325) | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Protein-ADP-ribose hydrolase, ... | | Authors: | Williams, R.S, Appel, C.D, Feld, G.K, Wallace, B.D. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the sirtuin-linked macrodomain SAV0325 from Staphylococcus aureus.
Protein Sci., 25, 2016
|
|
5L82
 
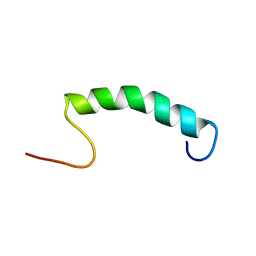 | |
5KJ5
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169D in complex with NAD+ | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
5KJD
 
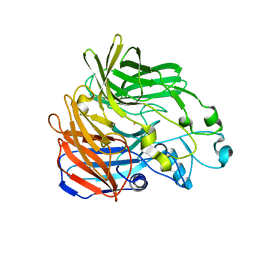 | |
5KLL
 
 | | Crystal structure of 2-hydroxymuconate-6-semialdehyde derived tautomeric intermediate in 2-aminomuconate 6-semialdehyde dehydrogenase N169D | | Descriptor: | (3~{E},5~{E})-6-oxidanyl-2-oxidanylidene-hexa-3,5-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KMD
 
 | | Structure of CavAb in complex with amlodipine | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-26 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
5KK0
 
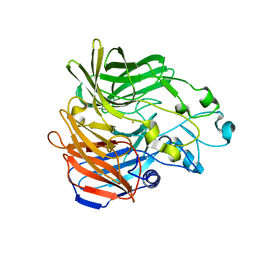 | | Synechocystis ACO mutant - T136A | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Utilization of Dioxygen by Carotenoid Cleavage Oxygenases.
J.Biol.Chem., 290, 2015
|
|
7Y9P
 
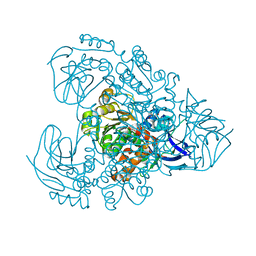 | | Xylitol dehydrogenase S96C/S99C/Y102C mutant(thermostabilized form) from Pichia stipitis | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular evolutionary insight of structural zinc atom in yeast xylitol dehydrogenases and its application in bioethanol production by lignocellulosic biomass.
Sci Rep, 13, 2023
|
|
7XXN
 
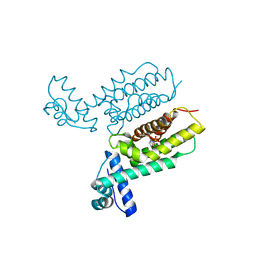 | | HapR Quadruple mutant, bound to Qstatin | | Descriptor: | 1-(5-bromanylthiophen-2-yl)sulfonylpyrazole, GLYCEROL, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY0
 
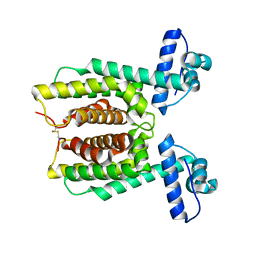 | | HapR Double mutant Y76F, F171C | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7Y4J
 
 | | HapR_Triple mutant Y76F, L97I, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXS
 
 | | HapR mutant I141V | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XYI
 
 | | HapR Quadruple mutant Y76F, L97I, I141V, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
