6JJ0
 
 | |
4XRF
 
 | |
1PSD
 
 | |
7E7V
 
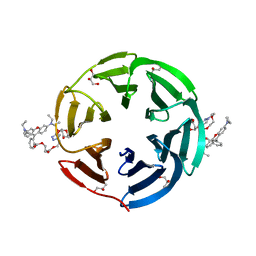 | | Crystal structure of RSL mutant in complex with sugar Ligand | | Descriptor: | 2-[2-[2-[4-[[(2R,3S,4R,5S,6S)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-1,2,3-triazol-1-yl]ethoxy]ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, Fucose-binding lectin protein,Fucose-binding lectin protein,Fucose-binding lectin protein, GLYCEROL | | Authors: | Li, L, Chen, G.S. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of RSL mutant in complex with Ligand
To Be Published
|
|
6UOJ
 
 | |
6UPM
 
 | |
7E7U
 
 | | Crystal structure of RSL mutant in complex with sugar Ligand | | Descriptor: | 2-[2-[4-[[(2R,3S,4R,5S,6S)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-1,2,3-triazol-1-yl]ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, Fucose-binding lectin protein,Fucose-binding lectin protein,Fucose-binding lectin protein, GLYCEROL | | Authors: | Li, L, Chen, G.S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RSL mutant in complex with Ligand
To Be Published
|
|
6UP9
 
 | |
6UPO
 
 | |
6UQ8
 
 | |
4P46
 
 | | J809.B5 Y31A TCR bound to IAb3K | | Descriptor: | 3K Peptide,H-2 class II histocompatibility antigen, A beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Stadinski, B.D, Huseby, E.S, Trenh, P, Stern, L.J. | | Deposit date: | 2014-03-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
6V61
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril.
To Be Published
|
|
3IRW
 
 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6V5M
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate.
To Be Published
|
|
6V71
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site
To Be Published
|
|
2HPG
 
 | |
3WMH
 
 | |
6JKE
 
 | | Discovery and the crystal structure of NS5 in complex with the N-terminal bromodomain of BRD2. | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, ... | | Authors: | Padmanabhan, B, Mathur, S, Deshmukh, P. | | Deposit date: | 2019-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel pyrano 1,3 oxazine based ligand inhibits the epigenetic reader hBRD2 in glioblastoma.
Biochem.J., 477, 2020
|
|
6V3T
 
 | |
2HRM
 
 | | Crystal structure of dUTPase complexed with substrate analogue methylene-dUTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-5'-O-[(S)-HYDROXY(PHOSPHONOMETHYL)PHOSPHORYL]URIDINE, Deoxyuridine 5'-triphosphate nucleotidohydrolase | | Authors: | Barabas, O, Kovari, J, Tapai, R, Vertessy, B.G. | | Deposit date: | 2006-07-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Methylene substitution at the alpha-beta bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site.
Proteins, 71, 2008
|
|
6C41
 
 | |
3ITZ
 
 | |
2CPY
 
 | | solution structure of RNA binding domain 3 in RNA binding motif protein 12 | | Descriptor: | RNA-binding protein 12 | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | solution structure of RNA binding domain 3 in RNA binding motif protein 12
To be Published
|
|
2CQG
 
 | | Solution structure of the RNA binding domain of TAR DNA-binding protein-43 | | Descriptor: | TAR DNA-binding protein-43 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of TAR DNA-binding protein-43
To be Published
|
|
4WSB
 
 | | Bat Influenza A polymerase with bound vRNA promoter | | Descriptor: | Influenza A polymerase vRNA promoter 3' end, Influenza A polymerase vRNA promoter 5' end, PHOSPHATE ION, ... | | Authors: | Cusack, S, Pflug, A, Guilligay, D, Reich, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
