2RLK
 
 | | Refined solution structure of porcine peptide YY (PYY) | | Descriptor: | Peptide YY | | Authors: | Neumoin, A, Mares, J, Lerch-Bader, M, Bader, R, Zerbe, O. | | Deposit date: | 2007-07-21 | | Release date: | 2007-08-14 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Probing the formation of stable tertiary structure in a model miniprotein at atomic resolution: determinants of stability of a helical hairpin
J.Am.Chem.Soc., 129, 2007
|
|
2RVQ
 
 | | Solution structure of the isolated histone H2A-H2B heterodimer | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J | | Authors: | Moriwaki, Y, Yamane, T, Ohtomo, H, Ikeguchi, M, Kurita, J, Sato, M, Nagadoi, A, Shimojo, H, Nishimura, Y. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the isolated histone H2A-H2B heterodimer
Sci Rep, 6, 2016
|
|
2VAI
 
 | | Solution structure of a B-DNA hairpin at high pressure | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural change in a B-DNA helix with hydrostatic pressure.
Nucleic Acids Res., 36, 2008
|
|
2VY4
 
 | | U11-48K CHHC ZN-FINGER DOMAIN | | Descriptor: | U11/U12 SMALL NUCLEAR RIBONUCLEOPROTEIN 48 KDA PROTEIN, ZINC ION | | Authors: | Tidow, H, Andreeva, A, Rutherford, T.J, Fersht, A.R. | | Deposit date: | 2008-07-17 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5' splice site of U12-type introns.
Structure, 17, 2009
|
|
2VCD
 
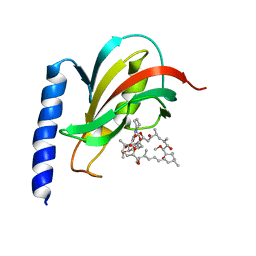 | | Solution structure of the FKBP-domain of Legionella pneumophila Mip in complex with rapamycin | | Descriptor: | Outer membrane protein MIP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Paschke, A.-K, Fischer, G, Roesch, P, Faber, C. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Legionella pneumophila Mip-rapamycin complex.
BMC Struct. Biol., 8, 2008
|
|
2VKC
 
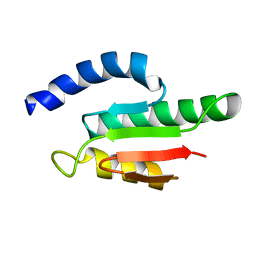 | | Solution structure of the B3BP Smr domain | | Descriptor: | NEDD4-BINDING PROTEIN 2 | | Authors: | Diercks, T, Ab, E, Daniels, M.A, deJong, R.N, Besseling, R, Kaptein, R, Folkers, G.E. | | Deposit date: | 2007-12-18 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the DNA- Binding Activity of the B3BP-Smr Domain.
J.Mol.Biol., 383, 2008
|
|
2WH9
 
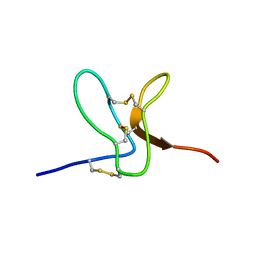 | | Solution structure of GxTX-1E | | Descriptor: | GUANGXITOXIN-1EGXTX-1E | | Authors: | Lee, S.K, Jung, H.H, Lee, J.Y, Lee, C.W, Kim, J.I. | | Deposit date: | 2009-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Gxtx-1E, a High Affinity Tarantula Toxin Interacting with Voltage Sensors in Kv2.1 Potassium Channels.
Biochemistry, 49, 2010
|
|
2WKS
 
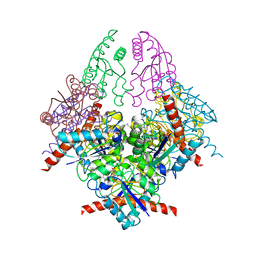 | | Structure of Helicobacter pylori Type II Dehydroquinase with a new carbasugar-thiophene inhibitor. | | Descriptor: | (1R,4S,5R)-1,4,5-trihydroxy-3-[(5-methyl-1-benzothiophen-2-yl)methoxy]cyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Guardado-Calvo, P, Llamas-Saiz, A.L, Prazeres, V.F.V, Tizon, L, Castedo, L, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues.
J. Med. Chem., 53, 2010
|
|
2V1S
 
 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
2W0P
 
 | |
2XJH
 
 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B, SODIUM ION | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
2VAH
 
 | | Solution structure of a B-DNA hairpin at low pressure. | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Change in a B-DNA Helix with Hydrostatic Pressure
Nucleic Acids Res., 36, 2008
|
|
2XJI
 
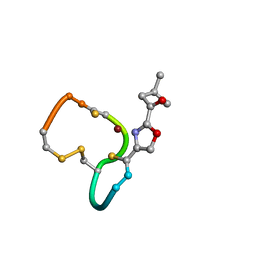 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
2V1T
 
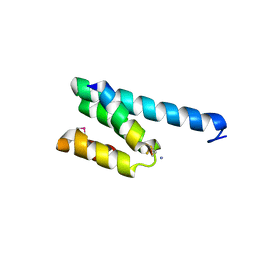 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
2VXE
 
 | |
2VY5
 
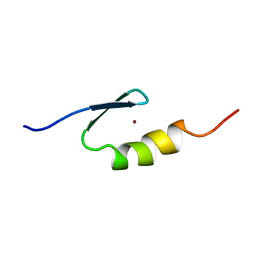 | | U11-48K CHHC Zn-finger protein domain | | Descriptor: | U11/U12 SMALL NUCLEAR RIBONUCLEOPROTEIN 48 KDA PROTEIN, ZINC ION | | Authors: | Tidow, H, Andreeva, A, Rutherford, T.J, Fersht, A.R. | | Deposit date: | 2008-07-18 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5' splice site of U12-type introns.
Structure, 17, 2009
|
|
2WXC
 
 | | The folding mechanism of BBL: Plasticity of transition-state structure observed within an ultrafast folding protein family. | | Descriptor: | DIHYDROLIPOYLTRANSSUCCINASE | | Authors: | Neuweiler, H, Sharpe, T.D, Rutherford, T.J, Johnson, C.M, Allen, M.D, Ferguson, N, Fersht, A.R. | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Folding Mechanism of Bbl: Plasticity of Transition-State Structure Observed within an Ultrafast Folding Protein Family.
J.Mol.Biol., 390, 2009
|
|
2WQG
 
 | | SAP domain from Tho1: L31W (fluorophore) mutant | | Descriptor: | PROTEIN THO1 | | Authors: | Dodson, C.A, Ferguson, N, Rutherford, T.J, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2009-08-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Engineering a Two-Helix Bundle Protein for Folding Studies.
Protein Eng.Des.Sel., 23, 2010
|
|
2XXS
 
 | | Solution structure of the N-terminal domain of the Shigella type III secretion protein MxiG | | Descriptor: | PROTEIN MXIG | | Authors: | McDowell, M.A, Johnson, S, Deane, J.E, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2010-11-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Studies on the N-Terminal Domain of the Shigella Type III Secretion Protein Mxig.
J.Biol.Chem., 286, 2011
|
|
2VXD
 
 | |
2Y4Q
 
 | |
2YKA
 
 | | RRM domain of mRNA export adaptor REF2-I bound to HVS ORF57 peptide | | Descriptor: | 52 KDA IMMEDIATE-EARLY PHOSPHOPROTEIN, RNA AND EXPORT FACTOR-BINDING PROTEIN 2 | | Authors: | Tunnicliffe, R.B, Hautbergue, G.M, Kalra, P, Wilson, S.A, Golovanov, A.P. | | Deposit date: | 2011-05-26 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri Orf57 to the Mammalian Export Adaptor Alyref.
Plos Pathog., 10, 2014
|
|
2N8A
 
 | | 1H, 13C and 15N chemical shift assignments and solution structure for PARP-1 F1F2 domains in complex with a DNA single-strand break | | Descriptor: | DNA (45-MER), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Wu, W. | | Deposit date: | 2015-10-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1.
Mol.Cell, 60, 2015
|
|
2MPM
 
 | | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: the N-terminal Region of CCR3 Bound to CCL11/Eotaxin-1 | | Descriptor: | CCR3, Eotaxin | | Authors: | Millard, C.J, Ludeman, J.P, Canals, M, Bridgford, J.L, Hinds, M.G, Clayton, D.J, Christopoulos, A, Payne, R.J, Stone, M.J. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-10 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: The N-Terminal Region of CCR3 Bound to CCL11/Eotaxin-1.
Structure, 22, 2014
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
