3WOA
 
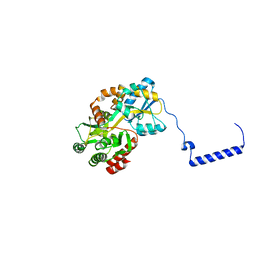 | |
8FU3
 
 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
8FMW
 
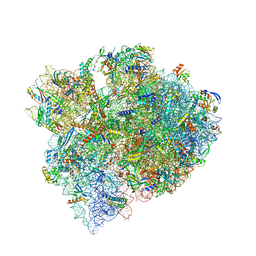 | | The structure of a hibernating ribosome in the Lyme disease pathogen | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Manjari, S.R, Agrawal, E.K, Keshavan, P, Koripella, R.K, Majumdar, S, Marcinkiewicz, A.L, Lin, Y.P, Agrawal, R.K, Banavali, N.K. | | Deposit date: | 2022-12-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | The structure of a hibernating ribosome in a Lyme disease pathogen.
Nat Commun, 14, 2023
|
|
8FN2
 
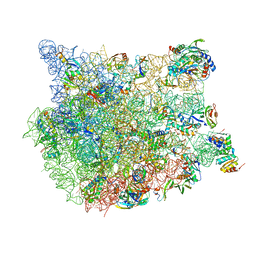 | | The structure of a 50S ribosomal subunit in the Lyme disease pathogen Borreliella burgdorferi | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Sharma, M.R, Manjari, S.R, Agrawal, E.K, Keshavan, P, Koripella, R.K, Majumdar, S, Marcinkiewicz, A.L, Lin, Y.P, Agrawal, R.K, Banavali, N.K. | | Deposit date: | 2022-12-26 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of a hibernating ribosome in a Lyme disease pathogen.
Nat Commun, 14, 2023
|
|
5CBN
 
 | | Fusion protein of mbp3-16 and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Maltose-binding periplasmic protein, mbp3-16,Immunoglobulin G-binding protein A | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-07-01 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
5CL1
 
 | | Complex structure of Norrin with human Frizzled 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Maltose-binding periplasmic protein,Norrin | | Authors: | Wang, Z, Ke, J, Shen, G, Cheng, Z, Xu, H.E, Xu, W. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
5C7R
 
 | | Revealing surface waters on an antifreeze protein by fusion protein crystallography | | Descriptor: | Fusion protein of Maltose-binding periplasmic protein and Type-3 ice-structuring protein HPLC 12, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sun, T, Gauthier, S, Campbell, R.L, Davies, P.L. | | Deposit date: | 2015-06-24 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Revealing Surface Waters on an Antifreeze Protein by Fusion Protein Crystallography Combined with Molecular Dynamic Simulations.
J.Phys.Chem.B, 119, 2015
|
|
8JBH
 
 | | Substance P bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq subunit alpha (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8JBG
 
 | | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
7A9C
 
 | |
8JI0
 
 | |
8IZJ
 
 | |
8G3W
 
 | | MBP-Mcl1 in complex with ligand 28 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3T
 
 | | MBP-Mcl1 in complex with ligand 12 | | Descriptor: | (1'S,3aS,5R,16R,17S,19Z,21R,21aR)-6'-chloro-20-fluoro-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-etheno-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3U
 
 | | MBP-Mcl1 in complex with ligand 21 | | Descriptor: | (1'S,3aS,5R,16R,17S,19Z,21R,21aR)-6'-chloro-20-fluoro-21-{[(5S,9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]methyl}-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-(ethanediylidene)-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, 1,2-ETHANEDIOL, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3S
 
 | | MBP-Mcl1 in complex with ligand 11 | | Descriptor: | (1'S,3aS,5R,16R,17S,19E,21S,21aR)-6'-chloro-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-etheno-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, FORMIC ACID, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3Y
 
 | | MBP-Mcl1 in complex with ligand 34 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3X
 
 | | MBP-Mcl1 in complex with ligand 32 | | Descriptor: | 1,2-ETHANEDIOL, Maltodextrin-binding protein, Induced myeloid leukemia cell differentiation protein Mcl-1 chimera, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8JA6
 
 | | Structure of the alginate epimerase/lyase complexed with tri-mannuronic acid | | Descriptor: | ACETATE ION, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, ... | | Authors: | Fujiwara, T. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the minimal bifunctional alginate epimerase AlgE3 from Azotobacter chroococcum.
Febs Lett., 598, 2024
|
|
8JAZ
 
 | | Structure of the alginate epimerase/lyase complexed with di-mannuronic acid | | Descriptor: | CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, mannuronan 5-epimerase | | Authors: | Fujiwara, T. | | Deposit date: | 2023-05-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis for the minimal bifunctional alginate epimerase AlgE3 from Azotobacter chroococcum.
Febs Lett., 598, 2024
|
|
8JA4
 
 | | Structure of the alginate epimerase/lyase | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Fujiwara, T. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis for the minimal bifunctional alginate epimerase AlgE3 from Azotobacter chroococcum.
Febs Lett., 598, 2024
|
|
8G8W
 
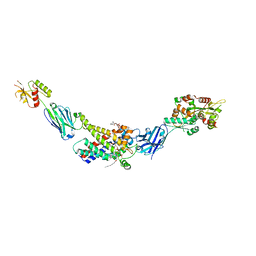 | | Molecular mechanism of nucleotide inhibition of human uncoupling protein 1 | | Descriptor: | CARDIOLIPIN, GUANOSINE-5'-TRIPHOSPHATE, Mitochondrial brown fat uncoupling protein 1, ... | | Authors: | Gogoi, P, Jones, S.A, Ruprecht, J.J, King, M.S, Lee, Y, Zogg, T, Pardon, E, Chand, D, Steimle, S, Copeman, D, Cotrim, C.A, Steyaert, J, Crichton, P.G, Moiseenkova-Bell, V, Kunji, E.R.S. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of purine nucleotide inhibition of human uncoupling protein 1.
Sci Adv, 9, 2023
|
|
4BL9
 
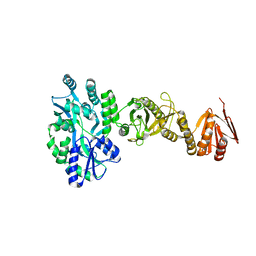 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLB
 
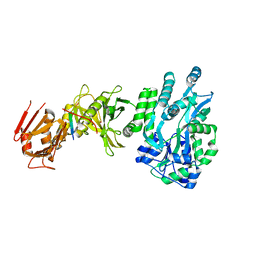 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI1p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, ZINC FINGER PROTEIN GLI1, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8GKH
 
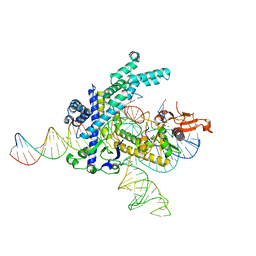 | | Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2023-03-19 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Fanzor is a eukaryotic programmable RNA-guided endonuclease.
Nature, 620, 2023
|
|
