1EFR
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT ALPHA, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT BETA, ... | | Authors: | Abrahams, J.P, Buchanan, S.K, Van Raaij, M.J, Fearnley, I.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-24 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of Bovine F1-ATPase Complexed with the Peptide Antibiotic Efrapeptin.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1P4S
 
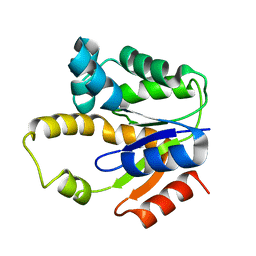 | |
2BWJ
 
 | | Structure of adenylate kinase 5 | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE 5, CHLORIDE ION, ... | | Authors: | Bunkoczi, G, Filippakopoulos, P, Fedorov, O, Jansson, A, Longman, E, Ugochukwu, E, Knapp, S, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Adenylate Kinase 5
To be Published
|
|
5ZZM
 
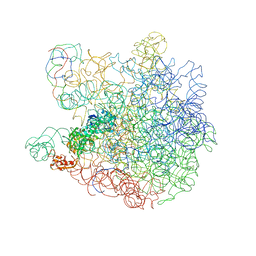 | |
3CW8
 
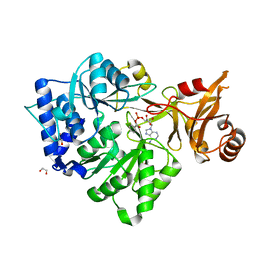 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, bound to 4CBA-Adenylate | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorobenzoyl CoA ligase, 5'-O-[(S)-{[(4-chlorophenyl)carbonyl]oxy}(hydroxy)phosphoryl]adenosine | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
3GIG
 
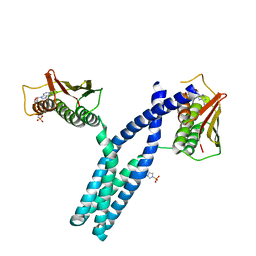 | | Crystal structure of phosphorylated DesKC in complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GIE
 
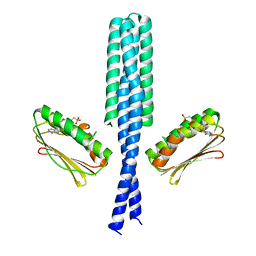 | | Crystal structure of DesKC_H188E in complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5IKO
 
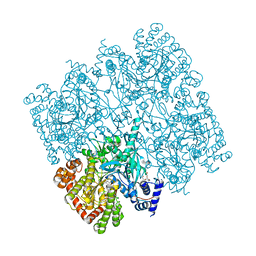 | | Crystal structure of human brain glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, brain form, HEXAETHYLENE GLYCOL, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
1FPB
 
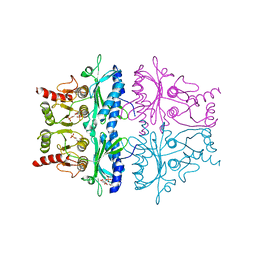 | | CRYSTAL STRUCTURE OF THE NEUTRAL FORM OF FRUCTOSE 1,6-BISPHOSPHATASE COMPLEXED WITH REGULATORY INHIBITOR FRUCTOSE 2,6-BISPHOSPHATE AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Liang, J.Y, Huang, S, Zhang, Y, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the neutral form of fructose 1,6-bisphosphatase complexed with regulatory inhibitor fructose 2,6-bisphosphate at 2.6-A resolution.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
8OXO
 
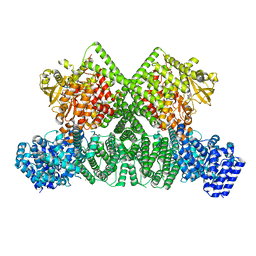 | | ATM(Q2971A) dimeric C-terminal region activated by oxidative stress in complex with Mg AMP-PNP and p53 peptide | | Descriptor: | Cellular tumor antigen p53, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Howes, A.C, Perisic, O, Williams, R.L. | | Deposit date: | 2023-05-02 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of ataxia-telangiectasia mutated by oxidative stress.
Sci Adv, 9, 2023
|
|
5K27
 
 | | Crystal structure of ancestral protein ancMT of ADP-dependent sugar kinases family. | | Descriptor: | ADENOSINE MONOPHOSPHATE, IODIDE ION, ancMT | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
1IXZ
 
 | | Crystal structure of the FtsH ATPase domain from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, MERCURY (II) ION, SULFATE ION | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IY1
 
 | | Crystal structure of the FtsH ATPase domain with ADP from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent metalloprotease FtsH | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IY2
 
 | | Crystal structure of the FtsH ATPase domain from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, SULFATE ION | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
8OXQ
 
 | | ATM(Q2971A) dimeric C-terminal region in complex with Mg AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine-protein kinase ATM, ... | | Authors: | Howes, A.C, Perisic, O, Williams, R.L. | | Deposit date: | 2023-05-02 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into the activation of ataxia-telangiectasia mutated by oxidative stress.
Sci Adv, 9, 2023
|
|
4YXR
 
 | | CRYSTAL STRUCTURE OF PKA IN COMPLEX WITH inhibitor. | | Descriptor: | 3-methyl-2H-indazole, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1MR2
 
 | | Structure of the MT-ADPRase in complex with 1 Mn2+ ion and AMP-CP (a inhibitor), a nudix enzyme | | Descriptor: | ADPR pyrophosphatase, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
2GDJ
 
 | | Delta-62 RADA recombinase in complex with AMP-PNP and magnesium | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, Y, Qian, X, He, Y, Luo, Y. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Rad51/RadA N-Terminal Domain Activates Nucleoprotein Filament ATPase Activity.
Structure, 14, 2006
|
|
1COW
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1ZXM
 
 | | Human Topo IIa ATPase/AMP-PNP | | Descriptor: | DNA topoisomerase II, alpha isozyme, MAGNESIUM ION, ... | | Authors: | Wei, H, Ruthenburg, A.J, Bechis, S.K, Verdine, G.L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Nucleotide-dependent Domain Movement in the ATPase Domain of a Human Type IIA DNA Topoisomerase.
J.Biol.Chem., 280, 2005
|
|
2GS7
 
 | | Crystal Structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
9BC7
 
 | | HCN1 M305L holo | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants.
Nature, 632, 2024
|
|
2DEI
 
 | | Crystal Structure of galaktokinase from Pyrococcus horikoshii with AMP-PNP and galactose | | Descriptor: | MAGNESIUM-5'-ADENYLY-IMIDO-TRIPHOSPHATE, Probable galactokinase, alpha-D-galactopyranose | | Authors: | Inagaki, E, Sakamoto, K, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-10 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of galaktokinase from Pyrococcus horikoshii
To be Published
|
|
2ZT8
 
 | | Crystal structure of human Glycyl-tRNA synthetase (GlyRS) in complex with Gly-AMP analog | | Descriptor: | Glycyl-tRNA synthetase, [(2S,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (aminoacetyl)sulfamate | | Authors: | Guo, R.T, Yang, X.L, Schimmel, P. | | Deposit date: | 2008-09-19 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structures and biochemical analyses suggest unique mechanism and role for human GlyRS in Ap4A homeostasis
To be Published
|
|
