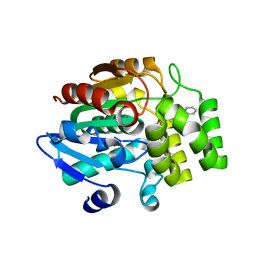
3WOA
 
 | |
8QAO
 
 | |
6FXA
 
 | |
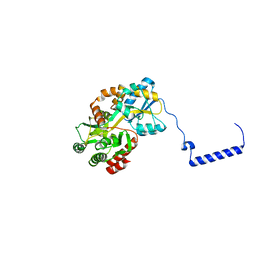
8EXR
 
 | | Cryo-EM structure of S. aureus BlaR1 TM and zinc metalloprotease domain | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Beta-lactam sensor/signal transducer BlaR1, PHOSPHATE ION, ... | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
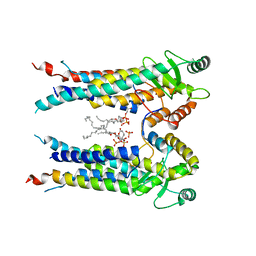
5DJ5
 
 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | Descriptor: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | Authors: | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
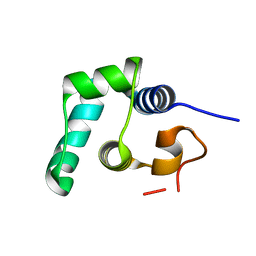
3ZHI
 
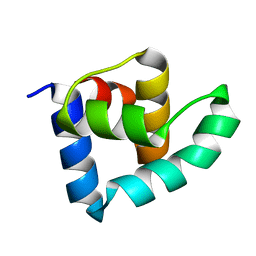 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 | | Descriptor: | CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
3ZHM
 
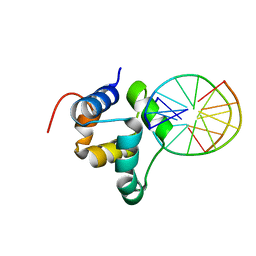 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 in complex with the OL2 operator half-site | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*AP*CP*TP*TP*GP*CP*AP*CP *TP*TP*GP*A)-3', 5'-D(*AP*GP*TP*TP*CP*AP*CP*GP*TP*TP*CP*AP*AP*GP *TP*GP*CP*A)-3', CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
2XRL
 
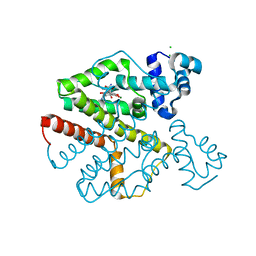 | | Tet-repressor class D T103A with doxycycline | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Palm, G.J, Waack, P, Hinrichs, W. | | Deposit date: | 2010-09-16 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
5D4Z
 
 | |
3E1K
 
 | |
1KCA
 
 | |
1R69
 
 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
3DZZ
 
 | |
2JFA
 
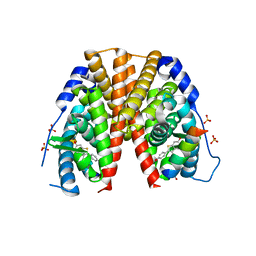 | | ESTROGEN RECEPTOR ALPHA LBD IN COMPLEX WITH AN AFFINITY-SELECTED COREPRESSOR PEPTIDE | | Descriptor: | COREPRESSOR PEPTIDE, ESTROGEN RECEPTOR, RALOXIFENE, ... | | Authors: | Heldring, N, Pawson, T, McDonnell, D, Treuter, E, Gustafsson, J.A, Pike, A.C.W. | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights Into Corepressor Recognition by Antagonist-Bound Estrogen Receptors.
J.Biol.Chem., 282, 2007
|
|
1PRU
 
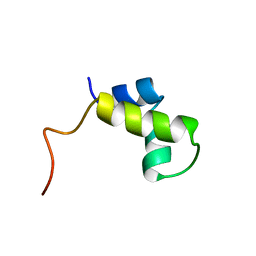 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
1PRV
 
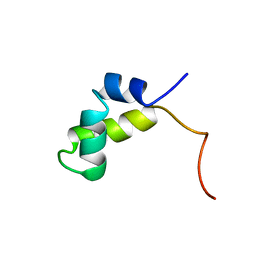 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
1JH9
 
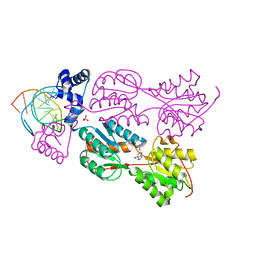 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PURF OPERATOR COMPLEX | | Descriptor: | 5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T)-3', HYPOXANTHINE, PHOSPHATE ION, ... | | Authors: | Huffman, J.L, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor.
Biochemistry, 41, 2002
|
|
1JHZ
 
 | | Purine Repressor Mutant Corepressor Binding Domain Structure | | Descriptor: | MAGNESIUM ION, PURINE NUCLEOTIDE SYNTHESIS REPRESSOR | | Authors: | Huffman, J.L, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 2001-06-28 | | Release date: | 2002-02-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor.
Biochemistry, 41, 2002
|
|
3WRP
 
 | |
3WG9
 
 | | Crystal structure of RSP, a Rex-family repressor | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
2VKE
 
 | |
3ERE
 
 | | Crystal structure of the arginine repressor protein from Mycobacterium tuberculosis in complex with the DNA operator | | Descriptor: | 5'-D(*DTP*DTP*DGP*DCP*DAP*DTP*DAP*DAP*DCP*DGP*DAP*DTP*DGP*DCP*DAP*DA)-3', 5'-D(*DTP*DTP*DGP*DCP*DAP*DTP*DCP*DGP*DTP*DTP*DAP*DTP*DGP*DCP*DAP*DA)-3', Arginine repressor, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the arginine repressor protein in complex with the DNA operator from Mycobacterium tuberculosis.
J.Mol.Biol., 384, 2008
|
|
6WJP
 
 | | Crystal structure of Arginine Repressor P115Q mutant from the pathogenic bacterium Corynebacterium pseudotuberculosis bound to arginine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ARGININE, ... | | Authors: | Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, de Morais, M.A.B, Murakami, M.T, Carareto, C.M.A, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A single P115Q mutation modulates specificity in the Corynebacterium pseudotuberculosis arginine repressor.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6WJO
 
 | | Crystal structure of wild-type Arginine Repressor from the pathogenic bacterium Corynebacterium pseudotuberculosis bound to tyrosine | | Descriptor: | Arginine repressor, SODIUM ION, SULFATE ION, ... | | Authors: | Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, de Morais, M.A.B, Murakami, M.T, Carareto, C.M.A, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | A single P115Q mutation modulates specificity in the Corynebacterium pseudotuberculosis arginine repressor.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
7OS9
 
 | | Crystal Structure of Domain Swapped Trp Repressor V58I Variant with purification tag | | Descriptor: | IMIDAZOLE, Trp operon repressor | | Authors: | Sprenger, J, Lawson, C.L, Lo Leggio, L, Von Wachenfeldt, C, Carey, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of Val58Ile tryptophan repressor in a domain-swapped array in the presence and absence of L-tryptophan.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
