2H96
 
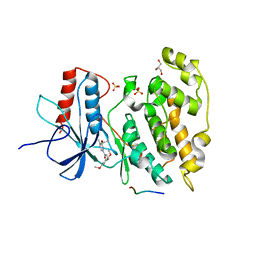 | | Discovery of Potent, Highly Selective, and Orally Bioavailable Pyridine Carboxamide C-jun NH2-terminal Kinase Inhibitors | | Descriptor: | 5-CYANO-N-(2,5-DIMETHOXYBENZYL)-6-ETHOXYPYRIDINE-2-CARBOXAMIDE, C-jun-amino-terminal kinase-interacting protein 1, GLYCEROL, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-06-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, highly selective, and orally bioavailable pyridine carboxamide c-Jun NH2-terminal kinase inhibitors.
J.Med.Chem., 49, 2006
|
|
2GMX
 
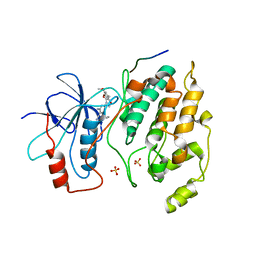 | | Selective Aminopyridine-Based C-Jun N-terminal Kinase inhibitors with cellular activity | | Descriptor: | C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, N-(4-AMINO-5-CYANO-6-ETHOXYPYRIDIN-2-YL)-2-(4-BROMO-2,5-DIMETHOXYPHENYL)ACETAMIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-04-07 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aminopyridine-Based c-Jun N-Terminal Kinase Inhibitors with Cellular Activity and Minimal Cross-Kinase Activity.
J.Med.Chem., 49, 2006
|
|
1EGP
 
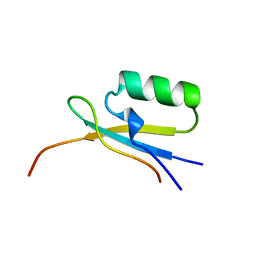 | |
1ELU
 
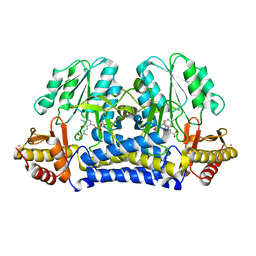 | | COMPLEX BETWEEN THE CYSTINE C-S LYASE C-DES AND ITS REACTION PRODUCT CYSTEINE PERSULFIDE. | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, ... | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E83
 
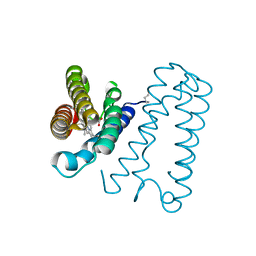 | | Cytochrome c' from Alcaligenes xylosoxidans - oxidized structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
4U01
 
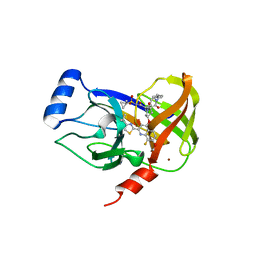 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3CX2
 
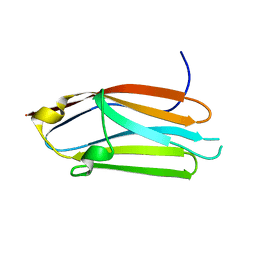 | | Crystal structure of the C1 domain of cardiac isoform of myosin binding protein-C at 1.3A | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Fisher, S.J, Helliwell, J.R, Khurshid, S, Govada, L, Redwood, C, Squire, J.M, Chayen, N.E. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An investigation into the protonation states of the C1 domain of cardiac myosin-binding protein C
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1KRJ
 
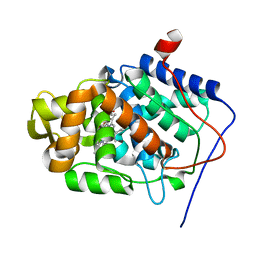 | | Engineering Calcium-binding site into Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome c Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Bhaskar, B, Sundaramoorthy, M, Poulos, T.L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conversion of an engineered potassium-binding site into a calcium-selective site in cytochrome c peroxidase.
J.Biol.Chem., 274, 1999
|
|
4C4J
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-{[2-chloro-4-(1-methyl-1H-imidazol-5-yl)phenyl]amino}-2-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
1KOK
 
 | | Crystal Structure of Mesopone Cytochrome c Peroxidase (MpCcP) | | Descriptor: | Cytochrome c Peroxidase, FE(III)-(4-MESOPORPHYRINONE) | | Authors: | Bhaskar, B, Immoos, C.E, Cohen, M.S, Barrows, T.P, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2001-12-20 | | Release date: | 2002-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mesopone cytochrome c peroxidase: functional model of heme oxygenated oxidases.
J.Inorg.Biochem., 91, 2002
|
|
5IP4
 
 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | Authors: | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
5O10
 
 | | Y48H mutant of human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Moreno-Chicano, T, Deacon, O.M, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Heightened Dynamics of the Oxidized Y48H Variant of Human Cytochrome c Increases Its Peroxidatic Activity.
Biochemistry, 56, 2017
|
|
8TAR
 
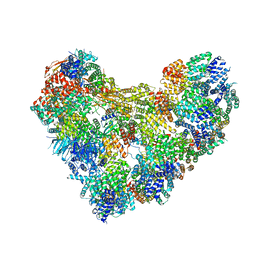 | | APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
8TAU
 
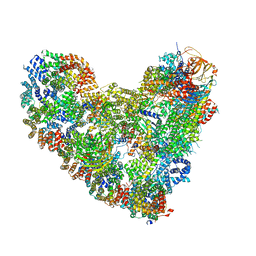 | | APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
8K3G
 
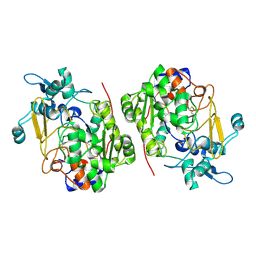 | |
9J8M
 
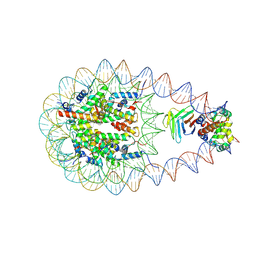 | | Cryo-EM structure of BAF-Lamin A/C IgF-nucleosome complex (High mobility complex) | | Descriptor: | Barrier-to-autointegration factor, DNA (193-MER), Histone H2A type 1-B/E, ... | | Authors: | Horikoshi, N, Miyake, R, Sogawa-Fujiwara, C, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Cryo-EM structures of the BAF-Lamin A/C complex bound to nucleosomes.
Nat Commun, 16, 2025
|
|
9J8O
 
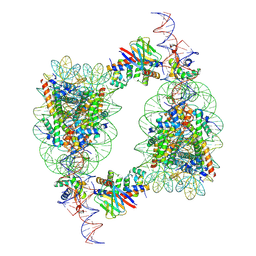 | | Cryo-EM structure of BAF-Lamin A/C IgF-H1-nucleosome complex | | Descriptor: | Barrier-to-autointegration factor, DNA (193-MER), Histone H1.1, ... | | Authors: | Horikoshi, N, Miyake, R, Sogawa-Fujiwara, C, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM structures of the BAF-Lamin A/C complex bound to nucleosomes.
Nat Commun, 16, 2025
|
|
9J8N
 
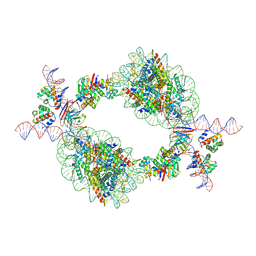 | | Cryo-EM structure of BAF-Lamin A/C IgF-nucleosome complex (Low mobility complex) | | Descriptor: | Barrier-to-autointegration factor, DNA (193-MER), Histone H2A type 1-B/E, ... | | Authors: | Horikoshi, N, Miyake, R, Sogawa-Fujiwara, C, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (7.14 Å) | | Cite: | Cryo-EM structures of the BAF-Lamin A/C complex bound to nucleosomes.
Nat Commun, 16, 2025
|
|
4WJY
 
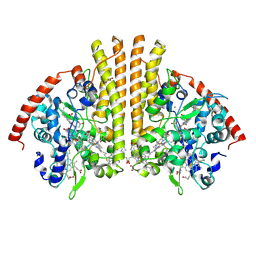 | | Esherichia coli nitrite reductase NrfA H264N | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Edwards, M.J, Lockwood, C.W.J. | | Deposit date: | 2014-10-01 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Resolution of key roles for the distal pocket histidine in cytochrome C nitrite reductases.
J.Am.Chem.Soc., 137, 2015
|
|
1F1F
 
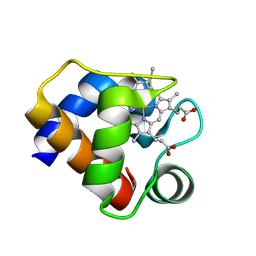 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
8OHM
 
 | |
8FSJ
 
 | | Cryo-EM structure of engineered hepatitis C virus E1E2 ectodomain in complex with antibodies AR4A, HEPC74, and IGH520 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AR4A heavy chain, ... | | Authors: | Metcalf, M.C, Ofek, G. | | Deposit date: | 2023-01-10 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.
Nat Commun, 14, 2023
|
|
1EB7
 
 | |
1EW3
 
 | | CRYSTAL STRUCTURE OF THE MAJOR HORSE ALLERGEN EQU C 1 | | Descriptor: | ALLERGEN EQU C 1 | | Authors: | Lascombe, M.B, Gregoire, C, Poncet, P, Tavares, G.A, Rosinski-Chupin, I, Rabillon, J, Goubran-Botros, H, Mazie, J.C, David, B, Alzari, P.M. | | Deposit date: | 2000-04-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the allergen Equ c 1. A dimeric lipocalin with restricted IgE-reactive epitopes.
J.Biol.Chem., 275, 2000
|
|
