8QLT
 
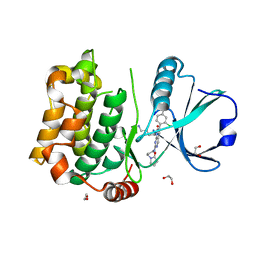 | | Human MST3 (STK24) kinase in complex with inhibitor MR30 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-[3-(2-oxidanylidenepyrrolidin-1-yl)propylamino]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8S7V
 
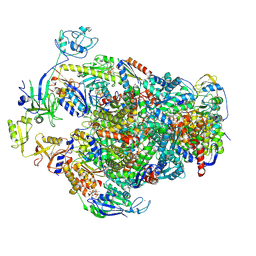 | | Methyl-coenzyme M reductase activation complex binding to the A2 component | | Descriptor: | 1-THIOETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Coenzyme B, ... | | Authors: | Ramirez-Amador, F, Paul, S, Kumar, A, Schuller, J.M. | | Deposit date: | 2024-03-04 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structure of the ATP-driven methyl-coenzyme M reductase activation complex.
Nature, 642, 2025
|
|
8S7X
 
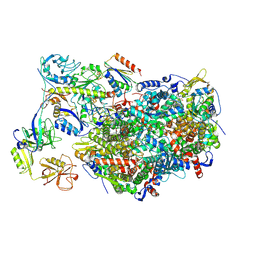 | | Methyl-coenzyme M reductase activation complex without the A2 component | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, DUF2098 domain-containing protein, ... | | Authors: | Ramirez-Amador, F, Paul, S, Kumar, A, Schuller, J.M. | | Deposit date: | 2024-03-04 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structure of the ATP-driven methyl-coenzyme M reductase activation complex.
Nature, 642, 2025
|
|
6G5A
 
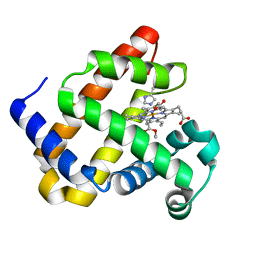 | | Heme-carbene complex in myoglobin H64V/V68A containing an N-methylhistidine as the proximal ligand, 1.48 angstrom resolution | | Descriptor: | ETHYL ACETATE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
6MUL
 
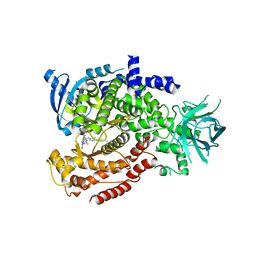 | | Murine PI3K delta kinsae domain - cpd 1 | | Descriptor: | 1-{1-[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]piperidin-4-yl}-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
8QCD
 
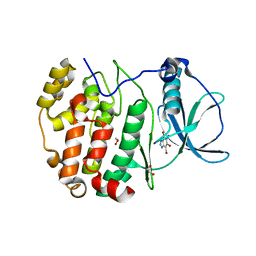 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR 4,5,6,7-TETRABROMOBENZOTRIAZOLE | | Descriptor: | 1,2-ETHANEDIOL, 4,5,6,7-TETRABROMOBENZOTRIAZOLE, Casein kinase II subunit alpha' | | Authors: | Werner, C, Niefind, K. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
6G5B
 
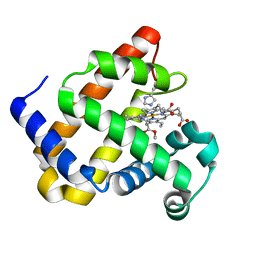 | | Heme-carbene complex in myoglobin H64V/V68A containing an N-methylhistidine as the proximal ligand, 1.6 angstrom resolution | | Descriptor: | ETHYL ACETATE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
8UOL
 
 | | Crystal structure of human NUAK1-MARK3 (6 mutations) kinase domain chimera bound with small molecule inhibitor #31 | | Descriptor: | 1,2-ETHANEDIOL, MAP/microtubule affinity-regulating kinase 3, Narazaciclib | | Authors: | Delker, S.L, Abendroth, J. | | Deposit date: | 2023-10-19 | | Release date: | 2024-10-23 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of UCB9386: A Potent, Selective, and Brain-Penetrant Nuak1 Inhibitor Suitable for In Vivo Pharmacological Studies.
J.Med.Chem., 67, 2024
|
|
8QBU
 
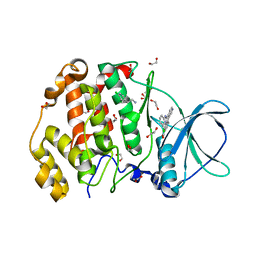 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR CX-4945 AND THE ALPHA-D-POCKET LIGAND 3,4-DICHLORO PHENETHYLAMINE (DPA) | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, ... | | Authors: | Werner, C, Niefind, K. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
4UWB
 
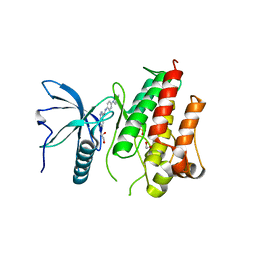 | | Fibroblast growth factor receptor 1 kinase in complex with JK-P5 | | Descriptor: | 1,2-ETHANEDIOL, FIBROBLAST GROWTH FACTOR RECEPTOR 1, N-[4-(4-methylpiperazin-1-yl)phenyl]-1H-indazole-3-carboxamide, ... | | Authors: | Beeston, H, Tucker, J, Kankanala, J. | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Validation of IMS-MS as a screening tool to identify type II kinase inhibitors of FGFR1 kinase.
Rapid Commun Mass Spectrom, 2021
|
|
6XI2
 
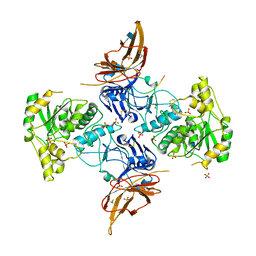 | | Apo form of POMGNT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-GLY-ALA-GLY-ALA-ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Halmo, S.M, Yeh, J, Wells, L, Moremen, K.W, Lanzilotta, W.N. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structures of beta-1,4-N-acetylglucosaminyltransferase 2: structural basis for inherited muscular dystrophies.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8YMN
 
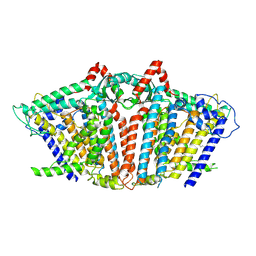 | | OSCA1.1-F516A pre-open 2 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Protein OSCA1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-03-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels.
Nat Commun, 15, 2024
|
|
6POJ
 
 | |
8V23
 
 | |
8YMQ
 
 | | OSCA1.1-F516A nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Protein OSCA1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-03-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels.
Nat Commun, 15, 2024
|
|
8YMM
 
 | | OSCA1.1-F516A open | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Protein OSCA1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-03-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels.
Nat Commun, 15, 2024
|
|
8YMP
 
 | | OSCA1.1-F516A nanodisc in LPC | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Protein OSCA1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-03-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels.
Nat Commun, 15, 2024
|
|
6G50
 
 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus as isolated. | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SULFATE ION, ... | | Authors: | Polyakov, K.M, Tsallagov, S.I, Tikhkonova, T.V, Popov, V.O. | | Deposit date: | 2018-03-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8QAO
 
 | |
8CKC
 
 | | Vitamin D receptor complex with 25-amine derivative of 1,25D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R)-6-azanyl-6-methyl-heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and biological activities of C25-amino and C25-nitro vitamin D analogs.
Bioorg.Chem., 136, 2023
|
|
8QUY
 
 | | Hexameric HIV-1 CA in complex with DDD01728501 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methylphenyl)methyl]-1~{H}-quinoxaline-2,3-dione, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QVA
 
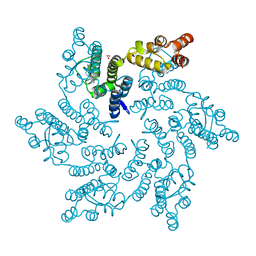 | | Hexameric HIV-1 CA in complex with DDD01829894 | | Descriptor: | 1,2-ETHANEDIOL, 7-azanyl-3-(phenylmethyl)-1~{H}-benzimidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8CK5
 
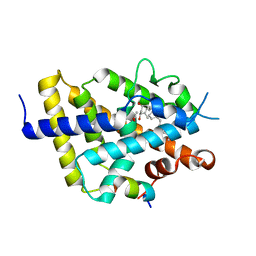 | | VDR LBD complex with 25-nitro derivative of 1,25D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-nitro-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and biological activities of C25-amino and C25-nitro vitamin D analogs.
Bioorg.Chem., 136, 2023
|
|
7ZKX
 
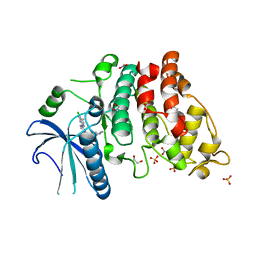 | | SRPK2 IN COMPLEX WITH INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-[3-[[[2-(6-chloranyl-5-fluoranyl-1H-benzimidazol-2-yl)pyrimidin-4-yl]amino]methyl]pyridin-2-yl]-N-methyl-methanesulfonamide, ... | | Authors: | Graedler, U. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | MSC-1186, a Highly Selective Pan-SRPK Inhibitor Based on an Exceptionally Decorated Benzimidazole-Pyrimidine Core.
J.Med.Chem., 66, 2023
|
|
8QUJ
 
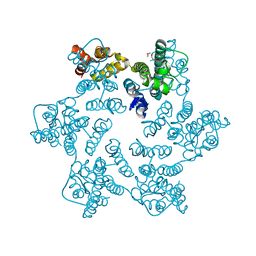 | | Hexameric HIV-1 CA in complex with DDD00100452 | | Descriptor: | 1,2-ETHANEDIOL, 3-(phenylmethyl)-1~{H}-imidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
