3IO2
 
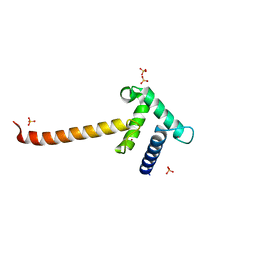 | | Crystal structure of the Taz2 domain of p300 | | Descriptor: | Histone acetyltransferase p300, SULFATE ION, ZINC ION | | Authors: | Miller, M, Dauter, Z, Wlodawer, A. | | Deposit date: | 2009-08-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Taz2 domain of p300: insights into ligand binding.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5V6Y
 
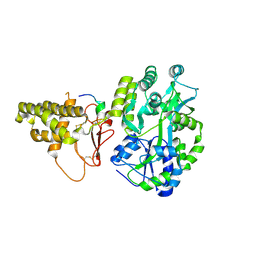 | |
1P2E
 
 | | H61A mutant of flavocytochrome c3 | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Rothery, E.L, Mowat, C.G, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2003-04-15 | | Release date: | 2003-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 61: An Important Heme Ligand in the Soluble Fumarate Reductase from Shewanella frigidimarina
Biochemistry, 42, 2003
|
|
8QNI
 
 | |
8QNH
 
 | | Crystal structure of the E3 ubiquitin ligase Cbl-b with an allosteric inhibitor (WO2020264398 Ex23) | | Descriptor: | 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Discovery of a Novel Benzodiazepine Series of Cbl-b Inhibitors for the Enhancement of Antitumor Immunity.
Acs Med.Chem.Lett., 14, 2023
|
|
1A08
 
 | |
1RY7
 
 | | Crystal Structure of the 3 Ig form of FGFR3c in complex with FGF1 | | Descriptor: | Fibroblast growth factor receptor 3, Heparin-binding growth factor 1 | | Authors: | Olsen, S.K, Ibrahimi, O.A, Raucci, A, Zhang, F, Eliseenkova, A.V, Yayon, A, Basilico, C, Linhardt, R.J, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5HVO
 
 | | Structure of Aspergillus fumigatus trehalose-6-phosphate synthase B in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase (UDP-forming), URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.467 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
2Y27
 
 | |
1A1B
 
 | |
2YMA
 
 | | X-ray structure of the Yos9 dimerization domain | | Descriptor: | PROTEIN OS-9 HOMOLOG | | Authors: | Hanna, J, Schuetz, A, Zimmermann, F, Behlke, J, Sommer, T, Heinemann, U. | | Deposit date: | 2011-06-07 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Structural and Biochemical Basis of Yos9 Protein Dimerization and Possible Contribution to Self-Association of 3-Hydroxy-3-Methylglutaryl-Coenzyme a Reductase Degradation Ubiquitin-Ligase Complex.
J.Biol.Chem., 287, 2012
|
|
1STS
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE FCHPQNT-NH2 DIMER | | Descriptor: | FCHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
1STR
 
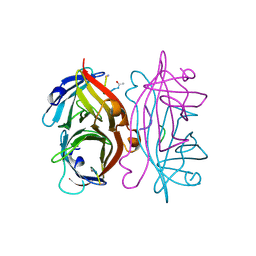 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE AC-CHPQNT-NH2 DIMER | | Descriptor: | AC-CHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
3QLW
 
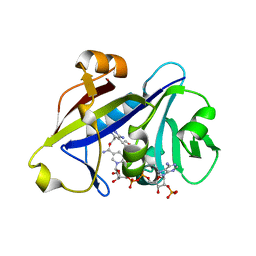 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein CaJ7.0360 | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
1A07
 
 | |
1A1C
 
 | |
3QLR
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine (UCP112A) | | Descriptor: | 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine, GLYCEROL, GLYCINE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
2X4N
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to residual fragments of a photocleavable peptide that is cleaved upon UV-light treatment | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
3N5A
 
 | | Synaptotagmin-7, C2B-domain, calcium bound | | Descriptor: | CALCIUM ION, Synaptotagmin-7 | | Authors: | Tomchick, D.R, Rizo, J, Craig, T.K. | | Deposit date: | 2010-05-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Structural and mutational analysis of functional differentiation between synaptotagmins-1 and -7.
Plos One, 5, 2010
|
|
4E8G
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound mg | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
6FI0
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with Fr 19 | | Descriptor: | 2-azanyl-1-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
2X4U
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 Peptide RT468-476 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
9D9Z
 
 | |
5BJV
 
 | | X-ray structure of the PglF UDP-N-acetylglucosamine 4,6-dehydratase from Campylobacterjejuni, D396N/K397A variant in complex with UDP-N-acrtylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5JXH
 
 | | Structure the proprotein convertase furin in complex with meta-guanidinomethyl-Phac-RVR-Amba at 2.0 Angstrom resolution. | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
