2PG1
 
 | |
8C18
 
 | | Solution structure of carotenoid-binding protein AstaPo1 in complex with astaxanthin | | Descriptor: | ASTAXANTHIN, Astaxanthin binding fasciclin family protein | | Authors: | Kornilov, F.D, Savitskaya, A.G, Slonimskiy, Y.B, Goncharuk, S.A, Sluchanko, N.N, Mineev, K.S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol, 6, 2023
|
|
4UVS
 
 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- pentyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-pentylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
6VQ7
 
 | |
8C0C
 
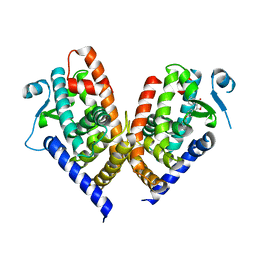 | | X-ray crystal structure of PPAR gamma ligand binding domain in complex with CZ46 | | Descriptor: | (2~{R})-2-[4-(naphthalen-1-ylmethoxy)phenyl]-4-oxidanyl-3-phenyl-2~{H}-furan-5-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Capelli, D, Montanari, R, Pochetti, G, Villa, S, Meneghetti, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biological Screening and Crystallographic Studies of Hydroxy gamma-Lactone Derivatives to Investigate PPAR gamma Phosphorylation Inhibition.
Biomolecules, 13, 2023
|
|
4UX4
 
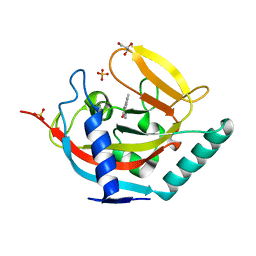 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-(4- methylphenyl)-5-oxo-5,6-dihydro-1,6-naphthyridin-1-ium | | Descriptor: | (1S)-1-methyl-7-(4-methylphenyl)-5-oxo-1,5-dihydro-1,6-naphthyridin-1-ium, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-19 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis and Evaluation in Vitro of Arylnaphthyridinones, Arylpyridopyrimidinones and Their Tetrahydro Derivatives as Inhibitors of the Tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
6VJJ
 
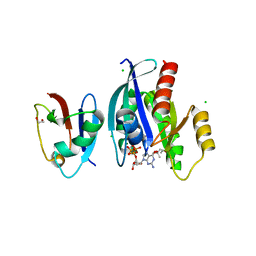 | | Crystal Structure of wild-type KRAS4b (GMPPNP-bound) in complex with RAS-binding domain (RBD) of RAF1/CRAF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
4V29
 
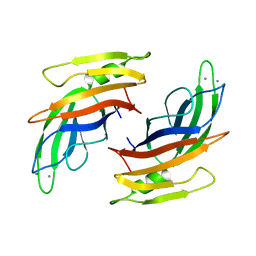 | |
2F4K
 
 | | Chicken villin subdomain HP-35, K65(NLE), N68H, K70(NLE), PH9 | | Descriptor: | Villin-1 | | Authors: | Chiu, T.K, Davies, D.R, Kubelka, J, Hofrichter, J, Eaton, W.A. | | Deposit date: | 2005-11-23 | | Release date: | 2006-04-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Sub-microsecond Protein Folding.
J.Mol.Biol., 359, 2006
|
|
6VQH
 
 | |
1M46
 
 | |
6VVF
 
 | |
8CH6
 
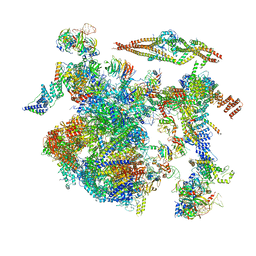 | | Structure of a late-stage activated spliceosome (BAqr) arrested with a dominant-negative Aquarius mutant (state B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Cretu, C, Schmitzova, J, Pena, V. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of catalytic activation in human splicing.
Nature, 617, 2023
|
|
6W13
 
 | |
2HBP
 
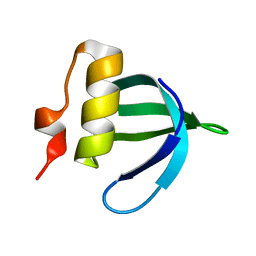 | | Solution Structure of Sla1 Homology Domain 1 | | Descriptor: | Cytoskeleton assembly control protein SLA1 | | Authors: | Overduin, M, Mahadev, R.K. | | Deposit date: | 2006-06-14 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Sla1p homology domain 1 and interaction with the NPFxD endocytic internalization motif.
Embo J., 26, 2007
|
|
4V8U
 
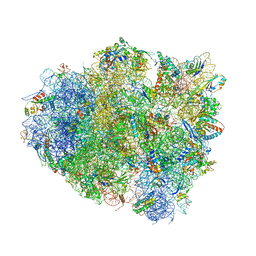 | | Crystal Structure of 70S Ribosome with Both Cognate tRNAs in the E and P Sites Representing an Authentic Elongation Complex. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.G, Feng, S, Chen, Y. | | Deposit date: | 2012-08-28 | | Release date: | 2014-07-09 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex.
PLoS ONE, 8, 2013
|
|
4W4K
 
 | |
8CJ3
 
 | | Urea-based foldamer inhibitor c3u_7 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_7 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ2
 
 | | Urea-based foldamer inhibitor c3u_5 chimera in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, SULFATE ION, ... | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ1
 
 | | Urea-based foldamer inhibitor c3u_3 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_3 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
4W6E
 
 | | Human Tankyrase 1 with small molecule inhibitor | | Descriptor: | 2-(4-{6-[(3S)-3,4-dimethylpiperazin-1-yl]-4-methylpyridin-3-yl}phenyl)-8-(hydroxymethyl)quinazolin-4(3H)-one, Tankyrase-1, ZINC ION | | Authors: | Kazmirski, S.L, Johannes, J, Boriack-Sjodin, P.A, Howard, T. | | Deposit date: | 2014-08-20 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology.
Acs Med.Chem.Lett., 6, 2015
|
|
4UVW
 
 | | Crystal structure of human tankyrase 2 in complex with 4,5-dimethyl-3- phenyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 4,5-dimethyl-3-phenylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
6W0M
 
 | | Human 8-oxoguanine glycosylase crosslinked with oxoG lesion containing DNA | | Descriptor: | 2-(2-ethoxyethoxy)ethanethiol, DNA (5'-D(P*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), ... | | Authors: | Shigdel, U, Verdine, G. | | Deposit date: | 2020-03-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The trajectory of intrahelical lesion recognition and extrusion by the human 8-oxoguanine DNA glycosylase.
Nat Commun, 11, 2020
|
|
6W0R
 
 | |
4UVY
 
 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-methoxy-1,2- dihydroisoquinolin-1-one | | Descriptor: | 3-(4-chlorophenyl)-5-methoxyisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
