5WPV
 
 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs at 3.59 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
1EYZ
 
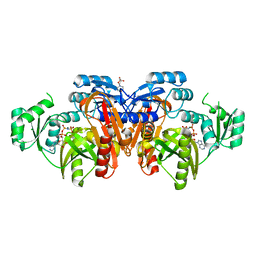 | | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG AND AMPPNP | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Firestine, S, Nixon, A, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2000-05-09 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
Biochemistry, 39, 2000
|
|
3UL5
 
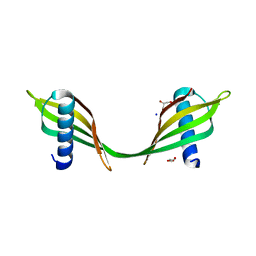 | | Saccharum officinarum canecystatin-1 in space group C2221 | | Descriptor: | Canecystatin-1, GLYCEROL, SODIUM ION | | Authors: | Valadares, N.F, Pereira, H.M, Oliveira-Silva, R, Garratt, R.C. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallography and NMR studies of domain-swapped canecystatin-1.
Febs J., 280, 2013
|
|
7D2A
 
 | | CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, AlyQ, CALCIUM ION, ... | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for binding uronic acids by family 32 carbohydrate-binding modules.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7CY1
 
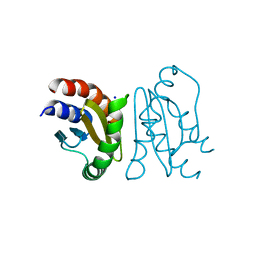 | | Crystal Structure of MglC from Myxococcus xanthus | | Descriptor: | Mutual gliding motility protein C, SODIUM ION | | Authors: | Thakur, K.G, Kapoor, S, Kodesia, A. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
J.Biol.Chem., 2021
|
|
5WKG
 
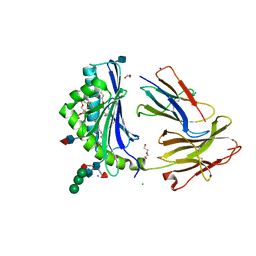 | | Crystal Structure of Human CD1b in Complex with PA | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shahine, A, Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A molecular basis of human T cell receptor autoreactivity toward self-phospholipids.
Sci Immunol, 2, 2017
|
|
7D3F
 
 | | Cryo-EM structure of human DUOX1-DUOXA1 in high-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dual oxidase 1, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
7D79
 
 | | The structure of DcsB complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DltD domain-containing protein, ... | | Authors: | Tang, Y, Zhou, J.H, Wang, G.Q. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.10411429 Å) | | Cite: | A Polyketide Cyclase That Forms Medium-Ring Lactones.
J.Am.Chem.Soc., 143, 2021
|
|
5WL1
 
 | | Crystal Structure of Human CD1b in Complex with PG | | Descriptor: | (19S,22R,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shahine, A, Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A molecular basis of human T cell receptor autoreactivity toward self-phospholipids.
Sci Immunol, 2, 2017
|
|
7D5E
 
 | | Left-handed G-quadruplex containing two bulges | | Descriptor: | 2xBulge-LHG4motif, POTASSIUM ION, SODIUM ION, ... | | Authors: | Das, P, Maity, A, Ngo, K.H, Winnerdy, F.R, Bakalar, B, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | Bulges in left-handed G-quadruplexes.
Nucleic Acids Res., 49, 2021
|
|
3VIH
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with glycerol | | Descriptor: | Beta-glucosidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1A50
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH 5-FLUOROINDOLE PROPANOL PHOSPHATE | | Descriptor: | 5-FLUOROINDOLE PROPANOL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
1B7O
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
7D10
 
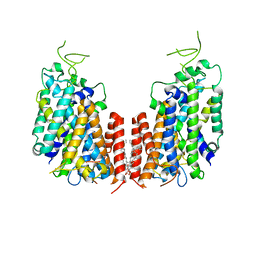 | | Human NKCC1 | | Descriptor: | PALMITIC ACID, Solute carrier family 12 member 2 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-12 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7D18
 
 | | Crystal structure of Acidobacteriales bacterium glutaminyl cyclase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Peptidase M28, ... | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
3VFA
 
 | | Crystal Structure of HIV-1 Protease Mutant V82A with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, SODIUM ION, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3VQJ
 
 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, ZINC ION | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-03-24 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
7D12
 
 | | NMR solution structures of CAG RNA-DB213 binding complex | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*CP*AP*GP*C)-3'), SODIUM ION | | Authors: | Chan, H.Y.E, Guo, P. | | Deposit date: | 2020-09-12 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CAG RNAs induce DNA damage and apoptosis by silencing NUDT16 expression in polyglutamine degeneration.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DPD
 
 | | Human MCM9 N-terminal domain | | Descriptor: | DNA helicase MCM9, SODIUM ION, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
5XAT
 
 | | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS | | Descriptor: | CITRATE ANION, Citrate-sodium symporter, SODIUM ION, ... | | Authors: | Jin, M.S, Kim, J.W, Kim, S, Kim, S, Lee, H, Lee, J.-O. | | Deposit date: | 2017-03-14 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS
Sci Rep, 7, 2017
|
|
7D2R
 
 | | Crystal structure of Agrobacterium tumefaciens aconitase X mutant - S449C/C510V | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, SODIUM ION, ... | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-09-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7CYF
 
 | | Cryo-EM structure of bicarbonate transporter SbtA in complex with PII-like signaling protein SbtB from Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION, ... | | Authors: | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-23 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
3VRK
 
 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase / Thiocyanate complex | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-04-11 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
7DCT
 
 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (24 bp) | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*AP*G)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
3VI2
 
 | | Crystal Structure Analysis of Plasmodium falciparum OMP Decarboxylase in complex with inhibitor HMOA | | Descriptor: | 4-(2-hydroxy-4-methoxyphenyl)-4-oxobutanoic acid, Orotidine 5'-phosphate decarboxylase, SODIUM ION | | Authors: | Takashima, Y, Mizohata, E, Krungkrai, S.R, Matsumura, H, Krungkrai, J, Horii, T, Inoue, T. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase
J.Biochem., 152, 2012
|
|
