5GLS
 
 | | Structure of bovine Lactoperoxidase with a partially modified covalent bond with heme moiety | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tiwari, P, Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution
Biochim. Biophys. Acta, 1865, 2016
|
|
4NKB
 
 | | Crystal Structure of the cryptic polo box (CPB)of ZYG-1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Shimanovskaya, E, Dong, G. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C. elegans ZYG-1 Cryptic Polo Box Suggests a Conserved Mechanism for Centriolar Docking of Plk4 Kinases.
Structure, 22, 2014
|
|
2AYL
 
 | | 2.0 Angstrom Crystal Structure of Manganese Protoporphyrin IX-reconstituted Ovine Prostaglandin H2 Synthase-1 Complexed With Flurbiprofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, GLYCEROL, ... | | Authors: | Gupta, K, Selinsky, B.S, Loll, P.J. | | Deposit date: | 2005-09-07 | | Release date: | 2006-01-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 angstroms structure of prostaglandin H2 synthase-1 reconstituted with a manganese porphyrin cofactor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7A9G
 
 | | Truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Mycobacterium tuberculosis with intermediate 2-acetyl-thiamine diphosphate | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, 2-ACETYL-THIAMINE DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-09-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First crystal structures of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis indicate a distinct mechanism of intermediate stabilization.
Sci Rep, 12, 2022
|
|
8U6T
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ758), a non-nucleoside inhibitor | | Descriptor: | 5-(2-{2-[2-oxo-3-(prop-2-enoyl)-2,3-dihydro-1H-benzimidazol-1-yl]ethoxy}phenoxy)naphthalene-2-carbonitrile, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
6RXC
 
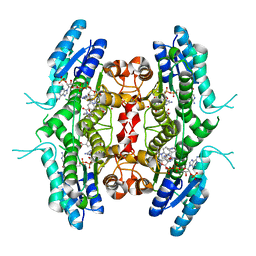 | | Leishmania major pteridine reductase 1 (LmPTR1) in complex with inhibitor 4 (NMT-C0026) | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1, methyl 1-[4-[[2,4-bis(azanyl)pteridin-6-yl]methyl-(3-oxidanylpropyl)amino]phenyl]carbonylpiperidine-4-carboxylate | | Authors: | Di Pisa, F, Dello Iacono, L, Pozzi, C, Mangani, S. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
J.Med.Chem., 65, 2022
|
|
4MSF
 
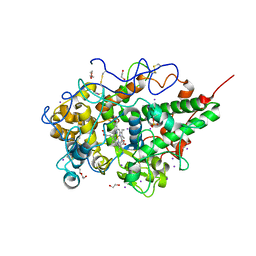 | | Crystal structure of the complex of goat lactoperoxidase with 3-hydroxymethyl phenol at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(hydroxymethyl)phenol, ... | | Authors: | Singh, A, Singh, R.P, Sinha, M, Singh, A.K, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with 3-hydroxymethyl phenol at 1.98 Angstrom resolution
To be published
|
|
5L42
 
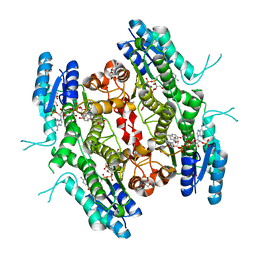 | | Leishmania major Pteridine reductase 1 (PTR1) in complex with compound 3 | | Descriptor: | (2~{R})-2-[3,4-bis(oxidanyl)phenyl]-6-oxidanyl-2,3-dihydrochromen-4-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Dello Iacono, L, Di Pisa, F, Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
6SZP
 
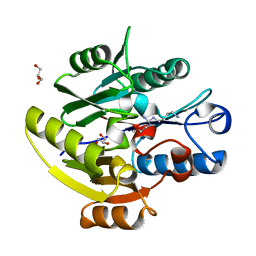 | | High resolution crystal structure of human DDAH-1 in complex with N-(4-Aminobutyl)-N'-(2-Methoxyethyl)guanidine | | Descriptor: | (1~{S})-~{N}'-(4-azanylbutyl)-~{N}"-(2-methoxyethyl)methanetriamine, GLYCEROL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Hennig, S, Vetter, I.R, Schade, D. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery ofN-(4-Aminobutyl)-N'-(2-methoxyethyl)guanidine as the First Selective, Nonamino Acid, Catalytic Site Inhibitor of Human Dimethylarginine Dimethylaminohydrolase-1 (hDDAH-1).
J.Med.Chem., 63, 2020
|
|
5F3M
 
 | | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complexed with L-neopterin at 1.5 Angstroms resolution . | | Descriptor: | 1,2-ETHANEDIOL, 7,8-dihydroneopterin aldolase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complexed with L-neopterin at 1.5 Angstroms resolution .
To Be Published
|
|
5I2G
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | Descriptor: | Diol dehydratase, S-1,2-PROPANEDIOL | | Authors: | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
6EBW
 
 | | hALK in complex with compound 9 (6-(((1S)-1-(5-Fluoropyridin-2-yl)ethyl)amino)-1-(3-methyl-1H-pyrazol-5-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl)(morpholin-4-yl)methanone | | Descriptor: | ALK tyrosine kinase receptor, [6-{[(1S)-1-(5-fluoropyridin-2-yl)ethyl]amino}-1-(5-methyl-1H-pyrazol-3-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl](morpholin-4-yl)methanone | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2018-08-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6EB2
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid | | Descriptor: | (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
5IU0
 
 | | Rubisco from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Valegaard, K, Hasse, D, Gunn, L, Andersson, I. | | Deposit date: | 2016-03-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure of Rubisco from Arabidopsis thaliana in complex with 2-carboxyarabinitol-1,5-bisphosphate.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6L2J
 
 | | Crystal structure of yak lactoperoxidase at 1.93 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Sharma, P, Rani, C, Sharma, S, Singh, T.P. | | Deposit date: | 2019-10-04 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Crystal structure of yak lactoperoxidase at 1.93 A resolution.
To Be Published
|
|
1BTO
 
 | |
9IZL
 
 | | hVanin-1 complexed with X17 | | Descriptor: | 8-oxa-2-azaspiro[4.5]decan-2-yl-[2-[(3~{S})-3-oxidanylpyrrolidin-1-yl]-1,3-thiazol-5-yl]methanone, Pantetheinase | | Authors: | Fan, S, Zhen, L, Xie, T. | | Deposit date: | 2024-08-01 | | Release date: | 2024-12-04 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of Thiazole Carboxamides as Novel Vanin-1 Inhibitors for Inflammatory Bowel Disease Treatment.
J.Med.Chem., 67, 2024
|
|
4O6V
 
 | |
6EB1
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid | | Descriptor: | (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
4OOF
 
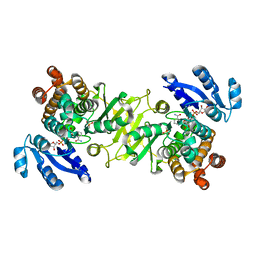 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203F mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
4OQM
 
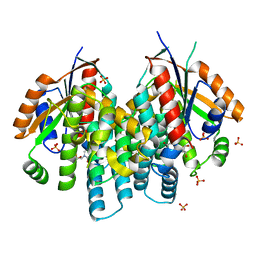 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with F-ARA-EdU | | Descriptor: | 1-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl)-5-ethynylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with F-ARA-EdU
To be Published
|
|
1A5Z
 
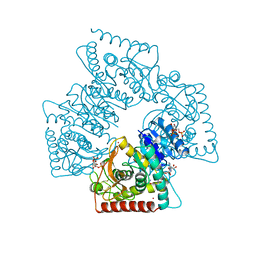 | | LACTATE DEHYDROGENASE FROM THERMOTOGA MARITIMA (TMLDH) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CADMIUM ION, L-LACTATE DEHYDROGENASE, ... | | Authors: | Auerbach, G, Ostendorp, R, Prade, L, Korndoerfer, I, Dams, T, Huber, R, Jaenicke, R. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization.
Structure, 6, 1998
|
|
6E0R
 
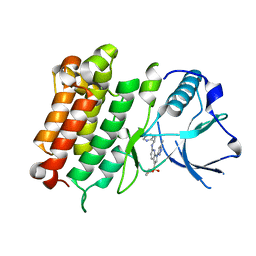 | | hALK in complex with compound 7 N-((1S)-1-(5-fluoropyridin-2-yl)ethyl)-1-(5-methyl-1H-pyrazol-3-yl)-3-(oxetan-3-ylsulfonyl)-1H-pyrrolo[2,3-b]pyridin-6-amine | | Descriptor: | ALK tyrosine kinase receptor, N-[(1S)-1-(5-fluoropyridin-2-yl)ethyl]-1-(5-methyl-1H-pyrazol-3-yl)-3-[(oxetan-3-yl)sulfonyl]-1H-pyrrolo[2,3-b]pyridin-6-amine | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2018-07-06 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6DB7
 
 | |
6DTP
 
 | |
