6BYW
 
 | | Structure of GoxA from Pseudoalteromonas luteoviolacea | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GoxA, ... | | Authors: | Yukl, E.T, Avalos, D. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-14 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Enzymatic Properties of an Unusual Cysteine Tryptophylquinone-Dependent Glycine Oxidase from Pseudoalteromonas luteoviolacea.
Biochemistry, 57, 2018
|
|
1RTL
 
 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN: NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND PYRROLIDINE-5,5-TRANSLACTAM INHIBITOR | | Descriptor: | N-[(2R,3S)-1-((2S)-2-{[(CYCLOPENTYLAMINO)CARBONYL]AMINO}-3-METHYLBUTANOYL)-2-(1-FORMYL-1-CYCLOBUTYL)PYRROLIDINYL]CYCLOPROPANECARBOXAMIDE, NS3 protease/helicase, NS4A COFACTOR, ... | | Authors: | Skarzynski, T, Somers, D.O.N. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 4. Incorporation of a P3/P4 urea leads to potent intracellular inhibitors of hepatitis C virus NS3/4A protease
Org.Lett., 5, 2003
|
|
1RPX
 
 | |
1QMY
 
 | | FMDV LEADER PROTEASE (LBSHORT-C51A-C133S) | | Descriptor: | 1,2-ETHANEDIOL, PROTEASE | | Authors: | Guarne, A, Tormo, J, Glaser, W, Skern, T, Fita, I. | | Deposit date: | 1999-10-08 | | Release date: | 2000-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Features Distinguish the Foot-and-Mouth Disease Virus Leader Proteinase from Other Papain-Like Enzymes
J.Mol.Biol., 302, 2000
|
|
6LLE
 
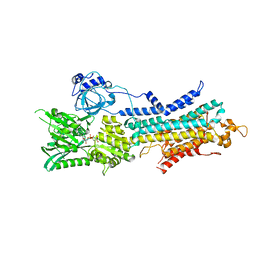 | | CryoEM structure of SERCA2b WT in E1-2Ca2+-AMPPCP state. | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
1LY4
 
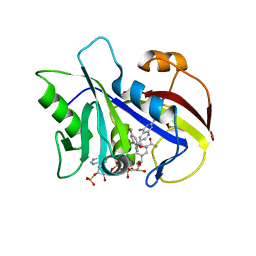 | | Analysis of quinazoline and PYRIDO[2,3D]PYRIMIDINE N9-C10 reversed bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3F6J
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-3)Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, F17a-G | | Authors: | Buts, L, de Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Biology, 2, 2013
|
|
1O4U
 
 | |
8UF5
 
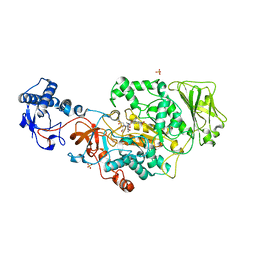 | | Catalytic domain of GtfB in complex with inhibitor G43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C, Velu, S. | | Deposit date: | 2023-10-03 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Discovery of Small Molecule Inhibitors of Cariogenic Virulence.
Sci Rep, 7, 2017
|
|
3I3S
 
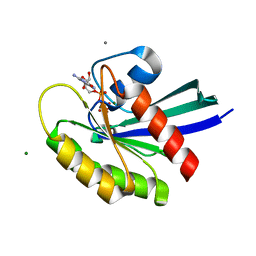 | | Crystal Structure of H-Ras with Thr50 replaced by Isoleucine | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Gremer, L, Dvorsky, R, Merbitz-Zahradnik, T, Wittinghofer, A, Ahmadian, M.R. | | Deposit date: | 2009-06-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A restricted spectrum of NRAS mutations causes Noonan syndrome.
Nat.Genet., 42, 2010
|
|
3HVG
 
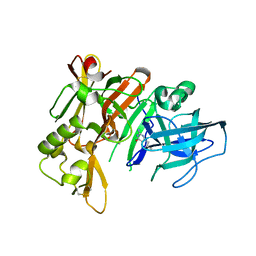 | | Structure of bace (beta secretase) in Complex with EV0 | | Descriptor: | 2-amino-6-propylpyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL | | Authors: | Godemann, R, Madden, J, Kramer, J, Smith, M.A, Barker, J, Ebneth, A. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Fragment-Based Discovery of BACE1 Inhibitors Using Functional Assays
Biochemistry, 48, 2009
|
|
3GNV
 
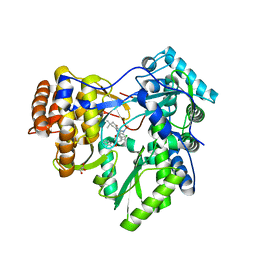 | | HCV NS5B polymerase in complex with 1,5 benzodiazepine inhibitor 1b | | Descriptor: | (11R)-10-acetyl-11-[4-(benzyloxy)-2-chlorophenyl]-6-hydroxy-3,3-dimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, GLYCEROL, RNA-directed RNA polymerase | | Authors: | De Bondt, H, Nyanguile, O. | | Deposit date: | 2009-03-18 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of a benzodiazepine scaffold yields a potent allosteric inhibitor of hepatitis C NS5B RNA polymerase.
J.Med.Chem., 52, 2009
|
|
3H5U
 
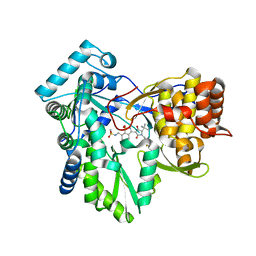 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor 1 | | Descriptor: | N-({3-[(5S)-5-tert-butyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzisothiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3GWE
 
 | |
6IMO
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[(1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
3H59
 
 | | Hepatitis C virus polymerase NS5B with thiazine inhibitor 2 | | Descriptor: | N-{3-[(5S)-5-(1,1-dimethylpropyl)-1-(4-fluoro-3-methylbenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-04-21 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 3: synthesis and optimization studies of benzothiazine-substituted tetramic acids
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1SHD
 
 | |
3H96
 
 | | Msmeg_3358 F420 Reductase | | Descriptor: | 1,2-ETHANEDIOL, F420-H2 Dependent Reductase A | | Authors: | Jackson, C.J, French, N, Newman, J, Taylor, M.C, Russell, R.J, Oakeshott, J.G. | | Deposit date: | 2009-04-30 | | Release date: | 2010-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of two families of F420 H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation.
Mol.Microbiol., 78, 2010
|
|
3HLN
 
 | | Crystal structure of ClpP A153C mutant with inter-heptamer disulfide bonds | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION | | Authors: | Kimber, M.S, Yu, A.Y.H, Borg, M, Chan, H.S, Houry, W.A. | | Deposit date: | 2009-05-27 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Theoretical Studies Indicate that the Cylindrical Protease ClpP Samples Extended and Compact Conformations.
Structure, 18, 2010
|
|
1MTN
 
 | | BOVINE ALPHA-CHYMOTRYPSIN:BPTI CRYSTALLIZATION | | Descriptor: | ALPHA-CHYMOTRYPSIN, BASIC PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Capasso, C, Rizzi, M, Menegatti, E, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-08-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the bovine alpha-chymotrypsin:Kunitz inhibitor complex. An example of multiple protein:protein recognition sites.
J.Mol.Recog., 10, 1997
|
|
3HW1
 
 | | Structure of Bace (beta secretase) in complex with ligand EV2 | | Descriptor: | 3-pyrrolidin-1-ylquinoxalin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Godemann, R, Madden, J, Kramer, J, Smith, M.A, Barker, J, Ebneth, A. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Fragment-Based Discovery of BACE1 Inhibitors Using Functional Assays
Biochemistry, 48, 2009
|
|
1O50
 
 | |
3FTP
 
 | |
3G4P
 
 | | OXA-24 beta-lactamase at pH 7.5 | | Descriptor: | Beta-lactamase OXA-24 | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2009-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase
J.Am.Chem.Soc., 132, 2010
|
|
1O5H
 
 | |
