2QBO
 
 | | Crystal structure of the P450cam G248V mutant in the cyanide bound state | | Descriptor: | CAMPHOR, CYANIDE ION, Cytochrome P450-cam, ... | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.D, Schlichting, I. | | Deposit date: | 2007-06-18 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alteration of P450 Distal Pocket Solvent Leads to Impaired Proton Delivery and Changes in Heme Geometry.
Biochemistry, 46, 2007
|
|
6L1F
 
 | |
3P1F
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with a hydroquinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-3,4-dihydroquinazolin-2(1H)-one, CREB-binding protein, ... | | Authors: | Filippakopoulos, P, Picaud, S, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with a hydroquinazolin ligand
To be Published
|
|
4E9C
 
 | | The structure of the polo-box domain (PBD) of polo-like kinase 1 (Plk1) in complex with LDPPLHSpTA phosphopeptide | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, GLYCEROL, LDPPLHSpTA phosphopeptide, ... | | Authors: | Sledz, P, Hyvonen, M, Lang, S, Stubbs, C.J, Abell, C. | | Deposit date: | 2012-03-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-throughput interrogation of ligand binding mode using a fluorescence-based assay.
Angew. Chem. Int. Ed. Engl., 51, 2012
|
|
3U1J
 
 | | Aprotinin bound to Dengue virus protease | | Descriptor: | Pancreatic trypsin inhibitor, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Noble, C.G. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand-bound structures of the dengue virus protease reveal the active conformation
J.Virol., 86, 2012
|
|
5BOJ
 
 | | Crystal Structure of Human Transthyretin in Complex with Gemfibrozil | | Descriptor: | 5-(2,5-dimethylphenoxy)-2,2-dimethylpentanoic acid, SODIUM ION, Transthyretin | | Authors: | Begum, A, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2015-05-27 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Enthalpic Forces Correlate with the Selectivity of Transthyretin-Stabilizing Ligands in Human Plasma.
J.Med.Chem., 58, 2015
|
|
6V84
 
 | |
1P2E
 
 | | H61A mutant of flavocytochrome c3 | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Rothery, E.L, Mowat, C.G, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2003-04-15 | | Release date: | 2003-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 61: An Important Heme Ligand in the Soluble Fumarate Reductase from Shewanella frigidimarina
Biochemistry, 42, 2003
|
|
1IF2
 
 | | X-RAY STRUCTURE OF LEISHMANIA MEXICANA TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH IPP | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE, [2(FORMYL-HYDROXY-AMINO)-ETHYL]-PHOSPHONIC ACID | | Authors: | Kursula, I, Partanen, S, Lambeir, A.-M, Antonov, D.M, Augustyns, K, Wierenga, R.K. | | Deposit date: | 2001-04-12 | | Release date: | 2001-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants for ligand binding and catalysis of triosephosphate isomerase.
Eur.J.Biochem., 268, 2001
|
|
4XDI
 
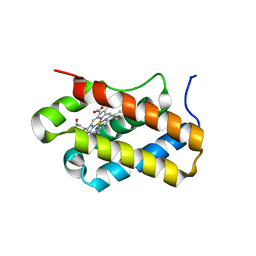 | | Structure of Chlamydomonas reinhardtii THB1 | | Descriptor: | Chlamydomonas reinhardtii THB1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rice, S.L, Boucher, L.E, Schlessman, J.L, Bosch, J, Lecomte, J.T.J. | | Deposit date: | 2014-12-19 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structure of Chlamydomonas reinhardtii THB1, a group 1 truncated hemoglobin with a rare histidine-lysine heme ligation.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4E8G
 
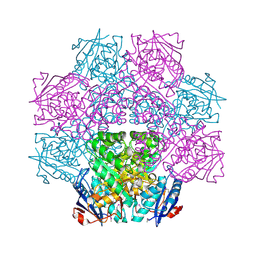 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound mg | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
5G1A
 
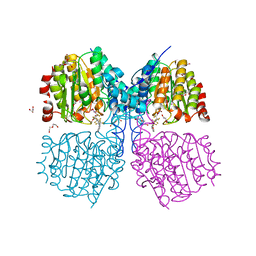 | | Bordetella Alcaligenes HDAH bound to PFSAHA | | Descriptor: | 2,2,3,3,4,4,5,5,6,6,7,7-dodecakis(fluoranyl)-~{N}-oxidanyl-~{N}'-phenyl-octanediamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
4OAL
 
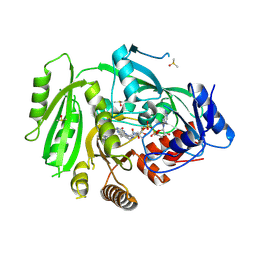 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor CPPU in alternative spacegroup | | Descriptor: | 1-(2-chloropyridin-4-yl)-3-phenylurea, Cytokinin dehydrogenase 4, DIMETHYL SULFOXIDE, ... | | Authors: | Kopecny, D, Morera, S, Vigouroux, A, Koncitikova, R. | | Deposit date: | 2014-01-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
6M65
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPNP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5G1B
 
 | | Bordetella Alcaligenes HDAH native | | Descriptor: | DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
6M69
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPCP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M6Y
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M72
 
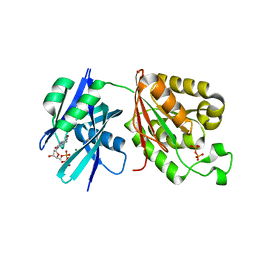 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGDP | | Descriptor: | 2'-deoxy-8-oxoguanosine 5'-(trihydrogen diphosphate), Hydrolase, NUDIX family protein, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1IEZ
 
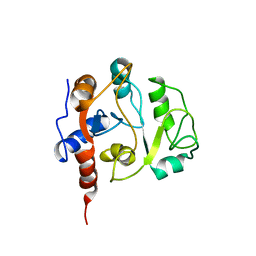 | | Solution Structure of 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase of Riboflavin Biosynthesis | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase | | Authors: | Kelly, M.J.S, Ball, L.J, Kuhne, R, Bacher, A, Oschkinat, H. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the 47-kDa dimeric enzyme 3,4-dihydroxy-2-butanone-4-phosphate synthase and ligand binding studies reveal the location of the active site.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3Q2F
 
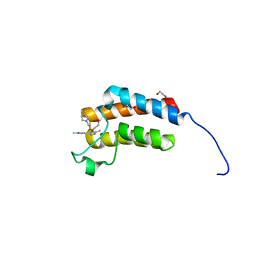 | | Crystal Structure of the bromodomain of human BAZ2B in complex with a triazolo ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[3-(1H-imidazol-1-yl)propyl]-5-methyl-4H-1,2,4-triazol-3-yl}-1-methyl-1H-pyrazol-5-amine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Muniz, J.R.C, Filippakopoulos, P, Picaud, S, Felletar, I, Keates, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the bromodomain of human BAZ2B in complex with a triazolo ligand
TO BE PUBLISHED
|
|
1U9E
 
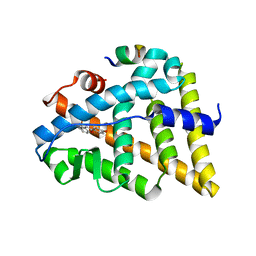 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-397 | | Descriptor: | 2-(4-HYDROXY-PHENYL)BENZOFURAN-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-09 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design Of Estrogen Receptor-beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
2Y4O
 
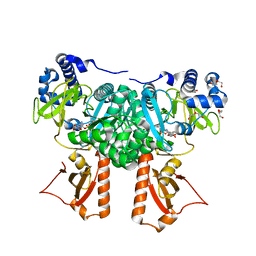 | | Crystal Structure of PaaK2 in complex with phenylacetyl adenylate | | Descriptor: | 5'-O-[HYDROXY(PHENYLACETYL)PHOSPHORYL]ADENOSINE, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Law, A, Boulanger, M.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Defining a Structural and Kinetic Rationale for Paralogous Copies of Phenylacetate-Coa Ligases from the Cystic Fibrosis Pathogen Burkholderia Cenocepacia J2315.
J.Biol.Chem., 286, 2011
|
|
6H1N
 
 | |
4P3K
 
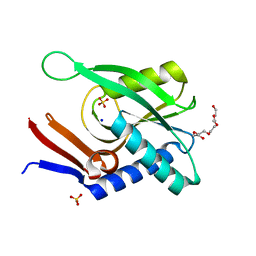 | | Structure of ancestral PyrR protein (PLUMPyrR) | | Descriptor: | Ancestral PyrR protein (Plum), PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P80
 
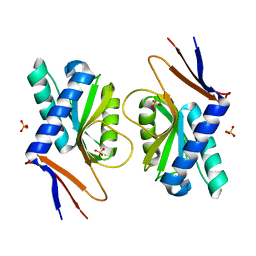 | | Structure of ancestral PyrR protein (AncGREENPyrR) | | Descriptor: | Ancestral PyrR protein (Green), SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-29 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
