2ZT6
 
 | |
2ZXF
 
 | |
1Q0Q
 
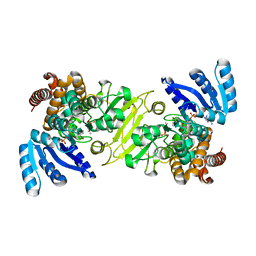 | | Crystal structure of DXR in complex with the substrate 1-deoxy-D-xylulose-5-phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-17 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
3FZG
 
 | | Structure of the 16S rRNA methylase ArmA | | Descriptor: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3MQ2
 
 | | Crystal Structure of 16S rRNA Methyltranferase KamB | | Descriptor: | 16S rRNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Macmaster, R.A. | | Deposit date: | 2010-04-27 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural insights into the function of aminoglycoside-resistance A1408 16S rRNA methyltransferases from antibiotic-producing and human pathogenic bacteria.
Nucleic Acids Res., 38, 2010
|
|
3HXW
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS in complex with SerSA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
3HXU
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS in complex with AlaSA | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
3HXV
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS in complex with GlySA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(glycylsulfamoyl)adenosine, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
3HXY
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS in complex with AMPPCP, Ala-AMP and PCP | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 5'-O-[(R)-{[(2S)-2-aminopropanoyl]oxy}(hydroxy)phosphoryl]adenosine, Alanyl-tRNA synthetase, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
3HY1
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS G237A in complex with SerSA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-HYDROXYETHYL DISULFIDE, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
3HXZ
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS G237A in complex with AlaSA | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
3HY0
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS G237A in complex with GlySA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(glycylsulfamoyl)adenosine, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
1TC5
 
 | |
1WNU
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with L-serine | | Descriptor: | SERINE, ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2004-08-09 | | Release date: | 2005-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WXO
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with zinc | | Descriptor: | ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2W53
 
 | | Structure of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. | | Descriptor: | REPRESSOR, SULFATE ION | | Authors: | Mate, M.J, Romero, A, Hernandez, A, Martinez, J.L. | | Deposit date: | 2008-12-04 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Smet, the Repressor of the Stenotrophomonas Maltophilia Multidrug Efflux Pump Smedef.
J.Biol.Chem., 284, 2009
|
|
4C13
 
 | | x-ray crystal structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ruane, K.M, Roper, D.I, Fulop, V, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy.
Nat.Chem.Biol., 2021
|
|
5K0M
 
 | | Targeting the PRC2 complex through a novel protein-protein interaction inhibitor of EED | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-{4-[4-(methylsulfonyl)piperazin-1-yl]phenyl}pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Bigelow, L.J, Zhu, H, Sun, C. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex.
Nat. Chem. Biol., 13, 2017
|
|
3C8Z
 
 | | The 1.6 A Crystal Structure of MshC: The Rate Limiting Enzyme in the Mycothiol Biosynthetic Pathway | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Cysteinyl-tRNA synthetase, ... | | Authors: | Tremblay, L.W, Fan, F, Vetting, M.W, Blanchard, J.S. | | Deposit date: | 2008-02-14 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of Mycobacterium smegmatis MshC: the penultimate enzyme in the mycothiol biosynthetic pathway.
Biochemistry, 47, 2008
|
|
2ADT
 
 | | NMR structure of a 30 kDa GAAA tetraloop-receptor complex. | | Descriptor: | 43-MER | | Authors: | Davis, J.H, Tonelli, M, Scott, L.G, Jaeger, L, Williamson, J.R, Butcher, S.E. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | RNA Helical Packing in Solution: NMR Structure of a 30 kDa GAAA Tetraloop-Receptor Complex
J.Mol.Biol., 351, 2005
|
|
2JXS
 
 | |
7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | Authors: | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
8R5I
 
 | | In situ structure of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions by flexible fitting into a cryoET map | | Descriptor: | Core protein A10, Core protein A4 | | Authors: | Calcraft, T, Hernandez-Gonzalez, M, Nans, A, Rosenthal, P.B, Way, M. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Palisade structure in intact vaccinia virions.
Mbio, 15, 2024
|
|
2YDH
 
 | |
2YGH
 
 | | SAM-I riboswitch with a G2nA mutation in the Kink turn in complex with S-adenosylmethionine | | Descriptor: | POTASSIUM ION, S-ADENOSYLMETHIONINE, SAM-I RIBOSWITCH, ... | | Authors: | Schroeder, K, Daldrop, P, Lilley, D. | | Deposit date: | 2011-04-17 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RNA Tertiary Interactions in a Riboswitch Stabilize the Structure of a Kink Turn.
Structure, 19, 2011
|
|
