5KA7
 
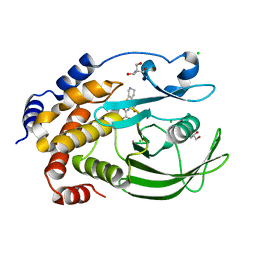 | | Protein Tyrosine Phosphatase 1B T178A mutant in complex with TCS401, closed state | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
3T8W
 
 | | A bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, N-((2R,3S,6S,18S,21S)-2-amino-18-(4-benzoylbenzyl)-21-carbamoyl-3-hydroxy-6-(naphthalen-2-ylmethyl)-4,7,16,19-tetraoxo-1-phenyl-11,14-dioxa-5,8,17,20-tetraazapentacosan-25-yl)hex-5-ynamide, ... | | Authors: | McGowan, S, Klemba, M, Greebaum, D.C. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4H44
 
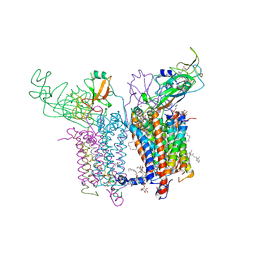 | | 2.70 A Cytochrome b6f Complex Structure From Nostoc PCC 7120 | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Hasan, S.S, Yamashita, E, Baniulis, D, Cramer, W.A. | | Deposit date: | 2012-09-16 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Quinone-dependent proton transfer pathways in the photosynthetic cytochrome b6f complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6WMT
 
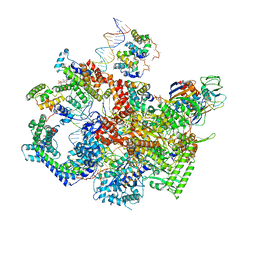 | | F. tularensis RNAPs70-(MglA-SspA)-ppGpp-PigR-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
5JWY
 
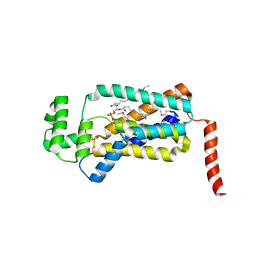 | | Structure of lipid phosphate phosphatase PgpB complex with PE | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Phosphatidylglycerophosphatase B | | Authors: | Tong, S, Wang, M, Zheng, L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-20 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insight into Substrate Selection and Catalysis of Lipid Phosphate Phosphatase PgpB in the Cell Membrane.
J.Biol.Chem., 291, 2016
|
|
3DPE
 
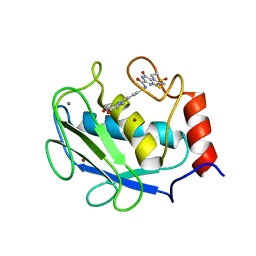 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | CALCIUM ION, N-{[2-(2-amino-3,4-dioxocyclobut-1-en-1-yl)-1,2,3,4-tetrahydroisoquinolin-7-yl]methyl}-4-oxo-3,5,6,8-tetrahydro-4H-thiopyrano[4',3':4,5]thieno[2,3-d]pyrimidine-2-carboxamide 7,7-dioxide, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
3KVW
 
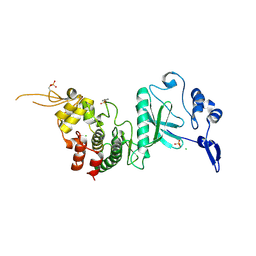 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand | | Descriptor: | (2Z,3E)-7'-bromo-3-(hydroxyimino)-2'-oxo-1,1',2',3-tetrahydro-2,3'-biindole-5-carboxylic acid, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Kritsanida, M, Magiatis, P, Skaltsounis, A.L, Soundararajan, M, Krojer, T, Gileadi, O, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand
To be Published
|
|
8YLB
 
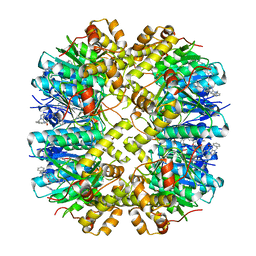 | | Cocrystal structures of agonists compound 1 with HsClpP | | Descriptor: | 5-[(2-methylphenyl)methyl]-11-(phenylmethyl)-2,5,7,11-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),6-dien-8-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Zhao, N, Zhu, Y, Bao, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational Design of a Novel Class of Human ClpP Agonists through a Ring-Opening Strategy with Enhanced Antileukemia Activity.
J.Med.Chem., 67, 2024
|
|
8BEL
 
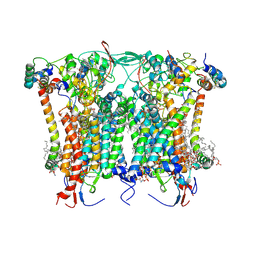 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII membrane domain) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
3G60
 
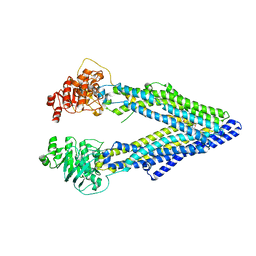 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | Descriptor: | (4R,11R,18R)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1a | | Authors: | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
5K0K
 
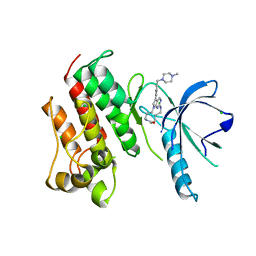 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2434 | | Descriptor: | 15-{4-[(4-methylpiperazin-1-yl)methyl]phenyl}-4,5,6,7,9,10,11,12-octahydro-2,16-(azenometheno)pyrrolo[2,1-d][1,3,5,9]te traazacyclotetradecin-8(3H)-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, X, Liu, J, Zhang, W, Stashko, M.A, Nichols, J, DeRyckere, D, Miley, M.J, Norris-Drouin, J, Chen, Z, Machius, M, Wood, E, Graham, D.K, Earp, H.S, Graham, K, Kireev, D, Frye, S.V. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Design and Synthesis of Novel Macrocyclic Mer Tyrosine Kinase Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
3BZ8
 
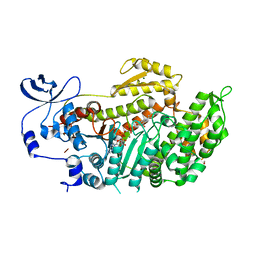 | | Crystal Structures of (S)-(-)-Blebbistatin Analogs bound to Dictyostelium discoideum myosin II | | Descriptor: | (3aS)-3a-hydroxy-7-methyl-1-phenyl-1,2,3,3a-tetrahydro-4H-pyrrolo[2,3-b]quinolin-4-one, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Allingham, J.S, Rayment, I. | | Deposit date: | 2008-01-17 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The small molecule tool (S)-(-)-blebbistatin: novel insights of relevance to myosin inhibitor design.
Org.Biomol.Chem., 6, 2008
|
|
3G5S
 
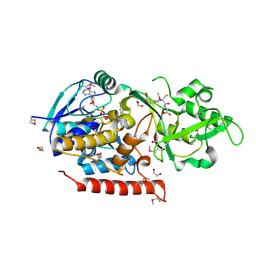 | | Crystal structure of Thermus thermophilus TrmFO in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4OGQ
 
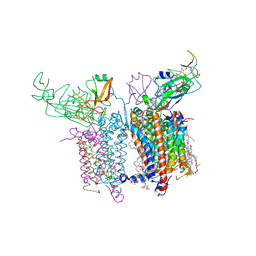 | | Internal Lipid Architecture of the Hetero-Oligomeric Cytochrome b6f Complex | | Descriptor: | (1R)-1-{[(2S)-3-hydroxy-2-{[(1R)-1-hydroxypentyl]oxy}propyl]oxy}hexan-1-ol, (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1S,8E)-1-{[(2S)-1-hydroxy-3-{[(1S)-1-hydroxypentadecyl]oxy}propan-2-yl]oxy}heptadec-8-en-1-ol, ... | | Authors: | Hasan, S.S, Cramer, W.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-05-14 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Internal Lipid Architecture of the Hetero-Oligomeric Cytochrome b6f Complex.
Structure, 22, 2014
|
|
5KAB
 
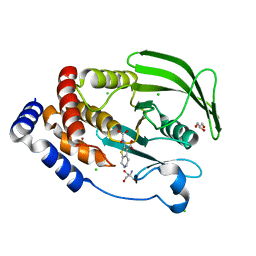 | | Protein Tyrosine Phosphatase 1B Delta helix 7, P185G mutant in complex with TCS401, open state | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Choy, M.S, Machado, L.E.S.F, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
3RIF
 
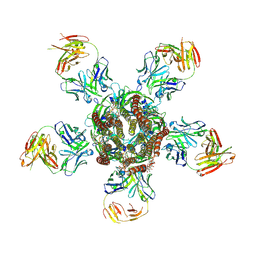 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and glutamate. | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avermectin-sensitive glutamate-gated chloride channel GluCl alpha, ... | | Authors: | Hibbs, R.E, Gouaux, E. | | Deposit date: | 2011-04-13 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | Principles of activation and permeation in an anion-selective Cys-loop receptor.
Nature, 474, 2011
|
|
5CEL
 
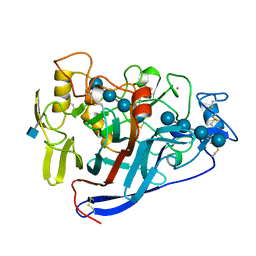 | | CBH1 (E212Q) CELLOTETRAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
5VYQ
 
 | | Crystal structure of the N-formyltransferase Rv3404c from mycobacterium tuberculosis in complex with YDP-Qui4N and folinic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dunsirn, M.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical Investigation of Rv3404c from Mycobacterium tuberculosis.
Biochemistry, 56, 2017
|
|
4CAW
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand | | Descriptor: | 2,6-DICHLORO-4-(2-PIPERAZIN-1-YLPYRIDIN-4-YL)-N-(1,5-DIMETHYL,3-ISOBUTYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Fang, W, Raimi, O.G, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
3OH3
 
 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE -Arabinose | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2S,3R,4S,5S)-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OFS
 
 | | Dynamic conformations of the CD38-mediated NAD cyclization captured using multi-copy crystallography | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2010-08-16 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Conformations of the CD38-Mediated NAD Cyclization Captured in a Single Crystal
J.Mol.Biol., 405, 2011
|
|
6CFD
 
 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
3L4T
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with BJ2661 | | Descriptor: | (1R,2S)-1-[(1S)-1,2-dihydroxyethyl]-3-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-2-hydroxypropyl sulfate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltase-glucoamylase, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
7F4S
 
 | | Crystal structure of TthMTA1-PteMTA9 complex | | Descriptor: | MT-a70 family protein, MTA9 | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
6JWV
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS A437G with STZ-DHP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 4-{[(2-amino-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}-N-(1,3-thiazol-2-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
