5DMR
 
 | |
6OA8
 
 | | Superfolder Green Fluorescent Protein with 4-cyano-L-phenylalanine at the chromophore (position 66) | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, SODIUM ION, ... | | Authors: | Piacentini, J, Olenginski, G.M, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
6YUH
 
 | | Crystal structure of SMYD3 with diperodon R enantiomer bound to allosteric site | | Descriptor: | Diperodon, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Cederfelt, D, Talibov, V.O, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase.
Chembiochem, 22, 2021
|
|
6XKI
 
 | | Crystal structure of eIF4A-I in complex with RNA bound to des-MePateA, a pateamine A analog | | Descriptor: | (3S,6Z,8E,11S,15R)-15-amino-3-[(1E,3E,5E)-7-(dimethylamino)-2,5-dimethylhepta-1,3,5-trien-1-yl]-9,11-dimethyl-4,12-dioxa-20-thia-21-azabicyclo[16.2.1]henicosa-1(21),6,8,18-tetraene-5,13-dione, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Liang, J, Naineni, S.K, Pelletier, J, Nagar, B. | | Deposit date: | 2020-06-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Functional mimicry revealed by the crystal structure of an eIF4A:RNA complex bound to the interfacial inhibitor, desmethyl pateamine A.
Cell Chem Biol, 28, 2021
|
|
6B9C
 
 | | Superfolder Green Fluorescent Protein with 4-nitro-L-phenylalanine at the chromophore (position 66) | | Descriptor: | CARBON DIOXIDE, Green fluorescent protein | | Authors: | Phillips-Piro, C.M, Brewer, S.H, Olenginski, G.M, Piacentini, J. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
6VJF
 
 | | The P-Loop K to A mutation of C. therm Vps1 GTPase-BSE | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Putative sorting protein Vps1 | | Authors: | Tornabene, B.A, Varlakhanova, N.V, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2020-01-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structural and functional characterization of the dominant negative P-loop lysine mutation in the dynamin superfamily protein Vps1.
Protein Sci., 29, 2020
|
|
5AEZ
 
 | |
5AEX
 
 | |
5AF1
 
 | |
6YN0
 
 | | Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity | | Descriptor: | Cell division protein FtsN, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Kerff, F, Terrak, M, Boes, A, Herman, H, Charlier, P. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The bacterial cell division protein fragment E FtsN binds to and activates the major peptidoglycan synthase PBP1b.
J.Biol.Chem., 295, 2020
|
|
8Q1T
 
 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with IOTA739 | | Descriptor: | 1,10-PHENANTHROLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Lund, B.A, Boronat, P, Hennig, S, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
7AAE
 
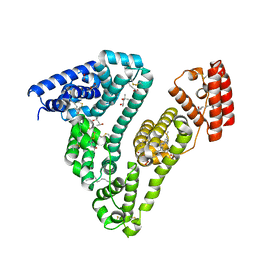 | | Crystal structure of Human serum albumin in complex with myristic acid at 2.27 Angstrom Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Albumin, FORMIC ACID, ... | | Authors: | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
7NDV
 
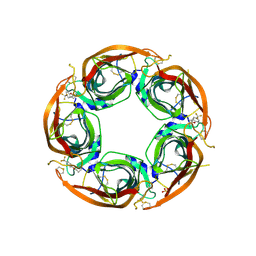 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001888. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[4-(trifluoromethyl)phenoxy]piperidine, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor
RSC Advances, 11, 2021
|
|
4XB4
 
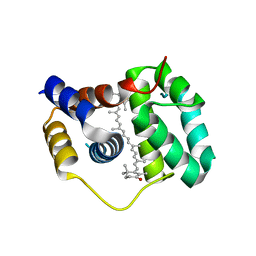 | | Structure of the N-terminal domain of OCP binding canthaxanthin | | Descriptor: | Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Kerfeld, C.A, Sutter, M, Leverenz, R.L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | PHOTOSYNTHESIS. A 12 angstrom carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection.
Science, 348, 2015
|
|
4XB5
 
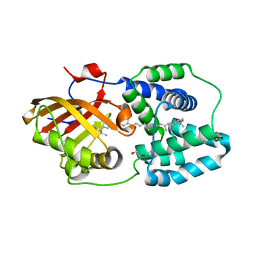 | | Structure of orange carotenoid protein binding canthaxanthin | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Kerfeld, C.A, Sutter, M, Leverenz, R.L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PHOTOSYNTHESIS. A 12 angstrom carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection.
Science, 348, 2015
|
|
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
1I92
 
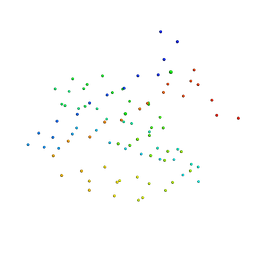 | | STRUCTURAL BASIS OF THE NHERF PDZ1-CFTR INTERACTION | | Descriptor: | CHLORIDE ION, NA+/H+ EXCHANGE REGULATORY CO-FACTOR | | Authors: | Karthikeyan, S, Leung, T, Ladias, J.A.A. | | Deposit date: | 2001-03-16 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Na+/H+ exchanger regulatory factor PDZ1 interaction with the carboxyl-terminal region of the cystic fibrosis transmembrane conductance regulator.
J.Biol.Chem., 276, 2001
|
|
3KF9
 
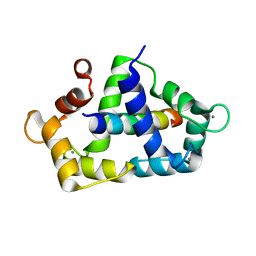 | | Crystal structure of the SdCen/skMLCK complex | | Descriptor: | CALCIUM ION, Caltractin, Myosin light chain kinase 2, ... | | Authors: | Radu, L, Assairi, L, Blouquit, Y, Durand, D, Miron, S, Charbonnier, J.B, Craescu, C.T. | | Deposit date: | 2009-10-27 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural features of the complexes formed by Scherffelia dubia centrin
To be Published
|
|
1M3J
 
 | |
1S70
 
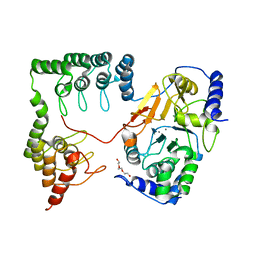 | | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) | | Descriptor: | 130 kDa myosin-binding subunit of smooth muscle myosin phophatase (M130), MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-beta (or delta) catalytic subunit, ... | | Authors: | Kerff, F, Terrak, M, Dominguez, R. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of protein phosphatase 1 regulation
Nature, 429, 2004
|
|
5N6O
 
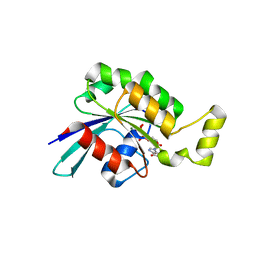 | | Wild type human Rac1-GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Cherfils, J, Ferrandez, Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Allosteric inhibition of the guanine nucleotide exchange factor DOCK5 by a small molecule.
Sci Rep, 7, 2017
|
|
1KDN
 
 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Cherfils, J, Xu, Y.W, Morera, S, Janin, J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2HP5
 
 | | Crystal Structure of the OXA-10 W154G mutant at pH 7.0 | | Descriptor: | Beta-lactamase PSE-2, COBALT (II) ION, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HP6
 
 | | Crystal structure of the OXA-10 W154A mutant at pH 7.5 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
