1KRN
 
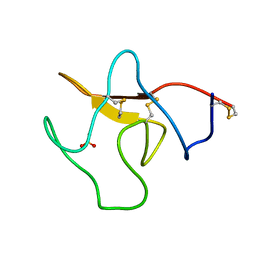 | | STRUCTURE OF KRINGLE 4 AT 4C TEMPERATURE AND 1.67 ANGSTROMS RESOLUTION | | Descriptor: | PLASMINOGEN, SULFATE ION | | Authors: | Stec, B, Teeter, M.M, Whitlow, M, Yamano, A. | | Deposit date: | 1995-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of human plasminogen kringle 4 at 1.68 a and 277 K. A possible structural role of disordered residues.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
8F74
 
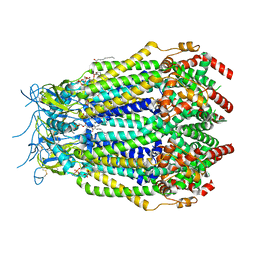 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1KXR
 
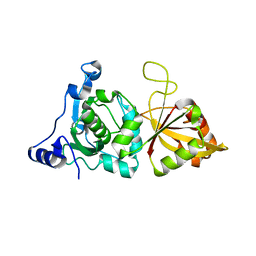 | | Crystal Structure of Calcium-Bound Protease Core of Calpain I | | Descriptor: | CALCIUM ION, thiol protease DOMAINS I AND II | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Elce, J.S, Jia, Z, Davies, P.L. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A Ca(2+) switch aligns the active site of calpain.
Cell(Cambridge,Mass.), 108, 2002
|
|
1KSQ
 
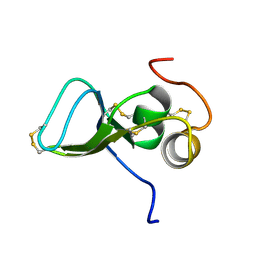 | | NMR Study of the Third TB Domain from Latent Transforming Growth Factor-beta Binding Protein-1 | | Descriptor: | LATENT TRANSFORMING GROWTH FACTOR BETA BINDING PROTEIN 1 | | Authors: | Lack, J, O'leary, J.M, Knott, V, Yuan, X, Rifkin, D.B, Handford, P.A, Downing, A.K. | | Deposit date: | 2002-01-14 | | Release date: | 2003-08-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Third TB Domain from LTBP1 Provides Insight into Assembly
of the Large Latent Complex that Sequesters Latent TGF-beta.
J.Mol.Biol., 334, 2003
|
|
1KJN
 
 | |
9F9Y
 
 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
1KTZ
 
 | | Crystal Structure of the Human TGF-beta Type II Receptor Extracellular Domain in Complex with TGF-beta3 | | Descriptor: | TGF-beta Type II Receptor, TRANSFORMING GROWTH FACTOR BETA 3 | | Authors: | Hart, P.J, Deep, S, Taylor, A.B, Shu, Z, Hinck, C.S, Hinck, A.P. | | Deposit date: | 2002-01-18 | | Release date: | 2002-02-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex.
Nat.Struct.Biol., 9, 2002
|
|
1KU3
 
 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment, Region 4 | | Descriptor: | sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
8F79
 
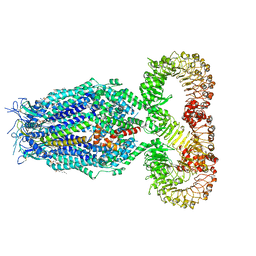 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F75
 
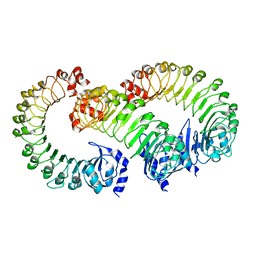 | | LRRC8A(T48D):C conformation 2 LRR focus | | Descriptor: | Volume-regulated anion channel subunit LRRC8A, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1KKH
 
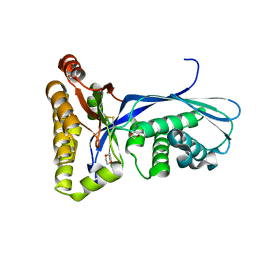 | | Crystal Structure of the Methanococcus jannaschii Mevalonate Kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate Kinase | | Authors: | Yang, D, Shipman, L.W, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2001-12-08 | | Release date: | 2002-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Methanococcus jannaschii mevalonate kinase, a member of the GHMP kinase superfamily.
J.Biol.Chem., 277, 2002
|
|
1KUW
 
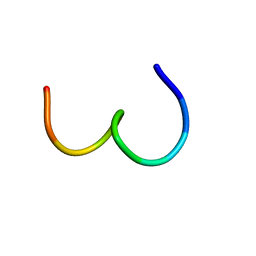 | | High-Resolution Structure and Localization of Amylin Nucleation Site in Detergent Micelles | | Descriptor: | Islet amyloid polypeptide | | Authors: | Mascioni, A, Porcelli, F, Ilangovan, U, Ramamoorthy, A, Veglia, G. | | Deposit date: | 2002-01-22 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences of the amylin nucleation site in SDS micelles: an NMR study.
Biopolymers, 69, 2003
|
|
5I5Y
 
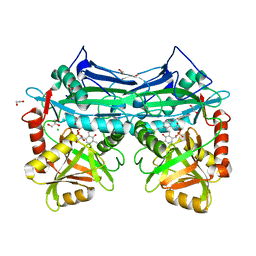 | |
1KVA
 
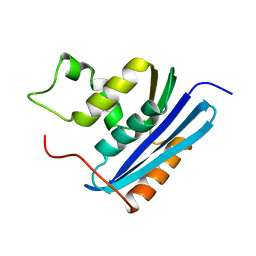 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVD
 
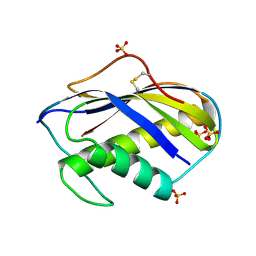 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | Descriptor: | SMK TOXIN, SULFATE ION | | Authors: | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
1KL5
 
 | |
1KM2
 
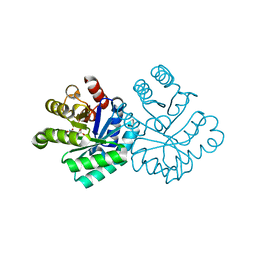 | |
4XOU
 
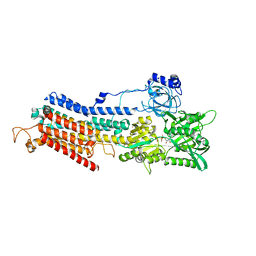 | | Crystal structure of the SR Ca2+-ATPase in the Ca2-E1-MgAMPPCP form determined by serial femtosecond crystallography using an X-ray free-electron laser. | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Nass, K, Drachmann, N.D, Markvardsen, A.J, Gutmann, M.J, Barends, T.R.M, Mattle, D, Shoeman, R.L, Doak, R.B, Boutet, S, Messerschmidt, M, Seibert, M.M, Williams, G.J, Foucar, L, Reinhard, L, Sitsel, O, Gregersen, J.L, Clausen, J.D, Boesen, T, Gotfryd, K, Wang, K.-T, Olesen, C, Moller, J.V, Nissen, P, Schlichting, I. | | Deposit date: | 2015-01-16 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of P-type ATPase-ligand complexes using an X-ray free-electron laser.
Iucrj, 2, 2015
|
|
1KM9
 
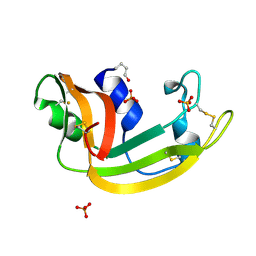 | | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog) | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE, OOCYTES | | Authors: | Chern, S.-S, Musayev, F.N, Amiraslanov, I.R, Liao, Y.-D, Liaw, Y.-C. | | Deposit date: | 2001-12-14 | | Release date: | 2003-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog)
To be Published
|
|
1KMJ
 
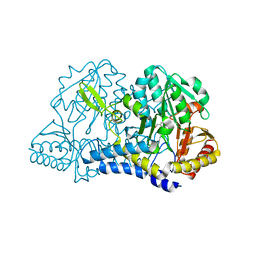 | |
1KMR
 
 | |
1KMX
 
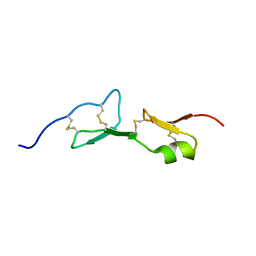 | |
4XPN
 
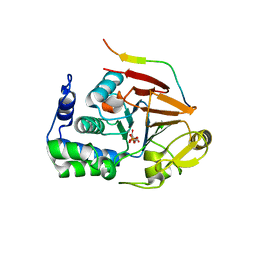 | | Crystal Structure of Protein Phosphate 1 complexed with PP1 binding domain of GADD34 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Structural and Functional Analysis of the GADD34:PP1 eIF2 alpha Phosphatase.
Cell Rep, 11, 2015
|
|
1KNQ
 
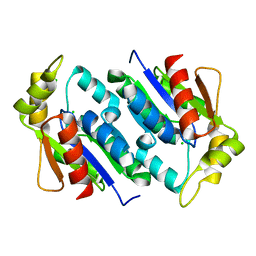 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KOQ
 
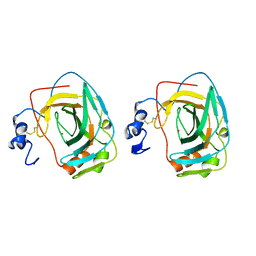 | | NEISSERIA GONORRHOEAE CARBONIC ANHYDRASE | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Huang, S, Xue, Y, Chirica, L, Lindskog, S, Jonsson, B.-H. | | Deposit date: | 1998-03-22 | | Release date: | 1998-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide.
J.Mol.Biol., 283, 1998
|
|
