5Z62
 
 | | Structure of human cytochrome c oxidase | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, COPPER (II) ION, ... | | Authors: | Gu, J, Zong, S, Wu, M, Yang, M. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-27 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the intact 14-subunit human cytochrome c oxidase.
Cell Res., 28, 2018
|
|
4KPR
 
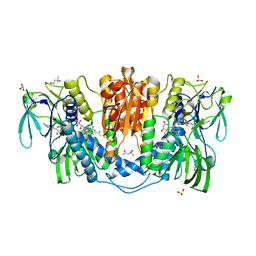 | | Tetrameric form of rat selenoprotein thioredoxin reductase 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, SULFITE ION, ... | | Authors: | Lindqvist, Y, Sandalova, T, Xu, J, Arner, E. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
To be Published, 2013
|
|
3X2G
 
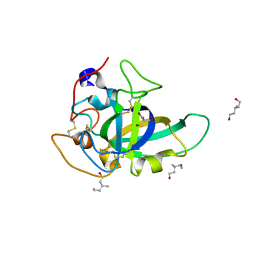 | | X-ray structure of PcCel45A N92D apo form at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
4HF3
 
 | |
3X2L
 
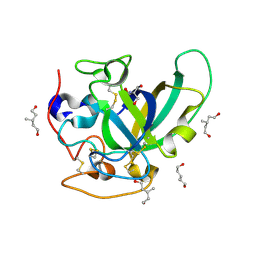 | | X-ray structure of PcCel45A apo form at 95K. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
2XAA
 
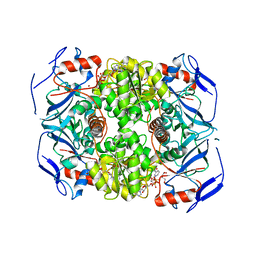 | | Alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541 at pH 8.5 in complex with NAD and butane-1,4-diol | | Descriptor: | 1,4-BUTANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SECONDARY ALCOHOL DEHYDROGENASE, ... | | Authors: | Kroutil, W, Gruber, K, Grogan, G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into Substrate Specificity and Solvent Tolerance in Alcohol Dehydrogenase Adh-'A' from Rhodococcus Ruber Dsm 44541.
Chem.Commun.(Camb.), 46, 2010
|
|
3N2B
 
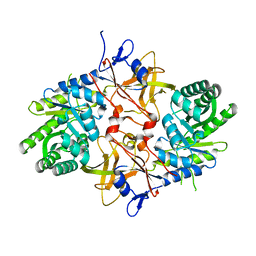 | | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae. | | Descriptor: | CHLORIDE ION, Diaminopimelate decarboxylase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae.
To be Published
|
|
3HS5
 
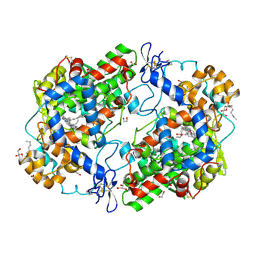 | | X-ray crystal structure of arachidonic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
2VZ0
 
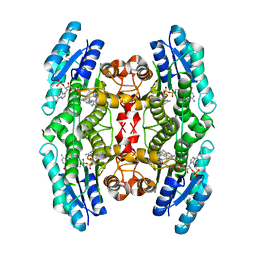 | | Pteridine Reductase 1 (PTR1) from Trypanosoma Brucei in complex with NADP and DDD00066641 | | Descriptor: | 6-(4-methylphenyl)quinazoline-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE | | Authors: | Robinson, D.A, Thompson, S, Sienkiewicz, N, Fairlamb, A.H. | | Deposit date: | 2008-07-29 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development and Validation of a Cytochrome C Coupled Assay for Pteridine Reductase 1 and Dihydrofolate Reductase.
Anal.Biochem., 396, 2010
|
|
1Z53
 
 | |
4GFS
 
 | | 1.8 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Nickel Bound at Active Site | | Descriptor: | 3-dehydroquinate dehydratase, NICKEL (II) ION, SUCCINIC ACID | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Nickel Bound at Active Site
TO BE PUBLISHED
|
|
3STH
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Toxoplasma gondii | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Staker, B.L, Edwards, T.E, Sankaran, B. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Membrane skeletal association and post-translational allosteric regulation of Toxoplasma gondii GAPDH1.
Mol.Microbiol., 103, 2017
|
|
3UUW
 
 | | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile.
TO BE PUBLISHED
|
|
3UH1
 
 | | Crystal Structure of Saccharopine Dehydrogenase from Saccharomyces cerevisiae with bound saccharopine and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID, ... | | Authors: | Kumar, V.P, Thomas, L.M, Bobyk, K.D, Andi, B, West, A.H, Cook, P.F. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
6D96
 
 | |
3HS7
 
 | | X-ray crystal structure of docosahexaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
4JA8
 
 | | Complex of Mitochondrial Isocitrate Dehydrogenase R140Q Mutant with AGI-6780 Inhibitor | | Descriptor: | 1-[5-(cyclopropylsulfamoyl)-2-thiophen-3-yl-phenyl]-3-[3-(trifluoromethyl)phenyl]urea, CALCIUM ION, GLYCEROL, ... | | Authors: | Wei, W, Chen, L, Wu, M, Jiang, F, Travins, J, Qian, K, DeLaBarre, B. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation.
Science, 340, 2013
|
|
1GLP
 
 | |
2Q1P
 
 | | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution | | Descriptor: | N-PROPANOL, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Kumar, S, Hariprasad, G, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-05-25 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution
To be Published
|
|
7D5W
 
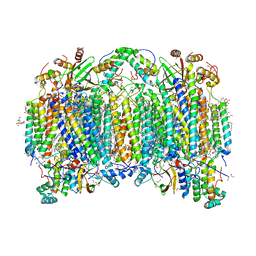 | | Bovine heart cytochrome c oxidase in a catalytic intermediate of O at 1.84 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2020-09-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Critical roles of the Cu B site in efficient proton pumping as revealed by crystal structures of mammalian cytochrome c oxidase catalytic intermediates.
J.Biol.Chem., 297, 2021
|
|
7D5X
 
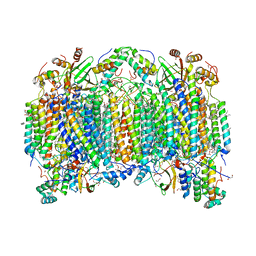 | | Bovine heart cytochrome c oxidase in a catalytic intermediate, IO10, at 1.74 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2020-09-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Critical roles of the Cu B site in efficient proton pumping as revealed by crystal structures of mammalian cytochrome c oxidase catalytic intermediates.
J.Biol.Chem., 297, 2021
|
|
1PHD
 
 | |
6MU3
 
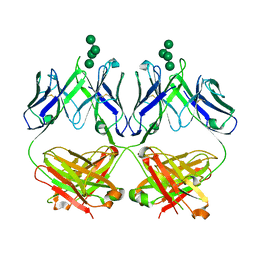 | | Anti-HIV-1 Fab 2G12 + Man7 re-refinement | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Wilson, I.A, Calarese, D.A, Stanfield, R.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6DDT
 
 | | mouse beta-mannosidase (MANBA) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gytz, H, Liang, J, Liang, Y, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2018-05-10 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of mammalian beta-mannosidase provides insight into beta-mannosidosis and nystagmus.
FEBS J., 286, 2019
|
|
6K9C
 
 | | The apo structure of NrS-1 C terminal region (305-718) | | Descriptor: | MERCURY (II) ION, Primase, SULFATE ION | | Authors: | Chen, X, Gan, J. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural studies reveal a ring-shaped architecture of deep-sea vent phage NrS-1 polymerase.
Nucleic Acids Res., 48, 2020
|
|
