1AGQ
 
 | | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR FROM RAT | | Descriptor: | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR | | Authors: | Eigenbrot, C, Gerber, N. | | Deposit date: | 1997-03-25 | | Release date: | 1997-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of glial cell-derived neurotrophic factor at 1.9 A resolution and implications for receptor binding.
Nat.Struct.Biol., 4, 1997
|
|
1A6G
 
 | | CARBONMONOXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-25 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
1ABW
 
 | | DEOXY RHB1.1 (RECOMBINANT HEMOGLOBIN) | | Descriptor: | HEMOGLOBIN-BASED BLOOD SUBSTITUTE, LEUCINE, METHIONINE, ... | | Authors: | Kundrot, C.E, Kroeger, K.S. | | Deposit date: | 1997-01-29 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a hemoglobin-based blood substitute: insights into the function of allosteric proteins.
Structure, 5, 1997
|
|
3K4V
 
 | | New crystal form of HIV-1 Protease/Saquinavir structure reveals carbamylation of N-terminal proline | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Olajuyigbe, F.M, Demitri, N, Ajele, J.O, Maurizio, E, Randaccio, L, Geremia, S. | | Deposit date: | 2009-10-06 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Carbamylation of N-terminal proline.
ACS Med Chem Lett, 1, 2010
|
|
1A3D
 
 | | PHOSPHOLIPASE A2 (PLA2) FROM NAJA NAJA VENOM | | Descriptor: | PHOSPHOLIPASE A2, SODIUM ION | | Authors: | Segelke, B.W, Nguyen, D, Chee, R, Xuong, H.N, Dennis, E.A. | | Deposit date: | 1998-01-20 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two novel crystal forms of Naja naja naja phospholipase A2 lacking Ca2+ reveal trimeric packing.
J.Mol.Biol., 279, 1998
|
|
2K62
 
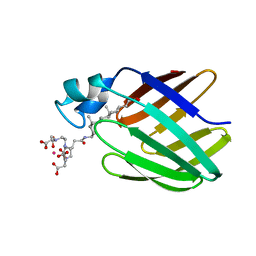 | | NMR solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based Gd(III)-chelate | | Descriptor: | (3alpha,5alpha,8alpha)-3-[(N,N-bis{2-[bis(carboxymethyl)amino]ethyl}-L-gamma-glutamyl)amino]cholan-24-oic acid, Liver fatty acid-binding protein, YTTERBIUM (III) ION | | Authors: | Tomaselli, S, Zanzoni, S, Ragona, L, Gianolio, E, Aime, S, Assfalg, M, Molinari, H. | | Deposit date: | 2008-07-03 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based gadolinium(III)-chelate, a potential hepatospecific magnetic resonance imaging contrast agent.
J.Med.Chem., 51, 2008
|
|
1ABQ
 
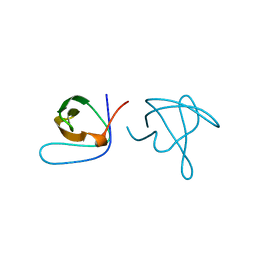 | |
7DD0
 
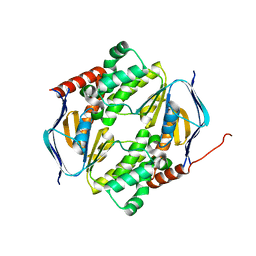 | |
1ALB
 
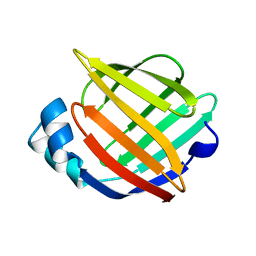 | |
2KHX
 
 | | Drosha double-stranded RNA binding motif | | Descriptor: | Ribonuclease 3 | | Authors: | Mueller, G.A, Miller, M, Ghosh, M, DeRose, E.F, London, R.E, Hall, T. | | Deposit date: | 2009-04-13 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Drosha double-stranded RNA-binding domain.
Silence, 1, 2010
|
|
3KA0
 
 | |
2KB6
 
 | | Solution structure of onconase C87A/C104A | | Descriptor: | Protein P-30 | | Authors: | Weininger, U, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Balbach, J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-11-24 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
1A01
 
 | | HEMOGLOBIN (VAL BETA1 MET, TRP BETA37 ALA) MUTANT | | Descriptor: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
1AJV
 
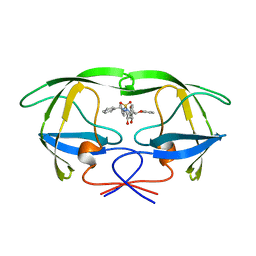 | | HIV-1 PROTEASE IN COMPLEX WITH THE CYCLIC SULFAMIDE INHIBITOR AHA006 | | Descriptor: | 2,7-DIBENZYL-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-[1,2,7]THIADIAZEPANE-4,5-DIOL, HIV-1 PROTEASE | | Authors: | Backbro, K, Unge, T. | | Deposit date: | 1997-05-11 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected binding mode of a cyclic sulfamide HIV-1 protease inhibitor.
J.Med.Chem., 40, 1997
|
|
2KNX
 
 | |
1A4R
 
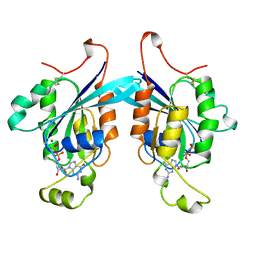 | | G12V MUTANT OF HUMAN PLACENTAL CDC42 GTPASE IN THE GDP FORM | | Descriptor: | AMINOPHOSPHONIC ACID-GUANYLATE ESTER, G25K GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rudolph, M.G, Vetter, I.R, Wittinghofer, A. | | Deposit date: | 1998-02-02 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleotide binding to the G12V-mutant of Cdc42 investigated by X-ray diffraction and fluorescence spectroscopy: two different nucleotide states in one crystal.
Protein Sci., 8, 1999
|
|
1A56
 
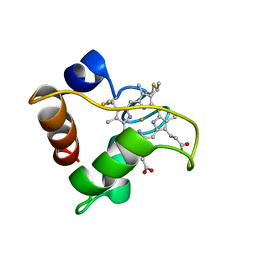 | | PRIMARY SEQUENCE AND SOLUTION CONFORMATION OF FERRICYTOCHROME C-552 FROM NITROSOMONAS EUROPAEA, NMR, MEAN STRUCTURE REFINED WITH EXPLICIT HYDROGEN BOND CONSTRAINTS | | Descriptor: | FERRICYTOCHROME C-552, HEME C | | Authors: | Timkovich, R, Bergmann, D, Arciero, D.M, Hooper, A.B. | | Deposit date: | 1998-02-20 | | Release date: | 1998-10-21 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Primary sequence and solution conformation of ferrocytochrome c-552 from Nitrosomonas europaea.
Biophys.J., 75, 1998
|
|
3JW1
 
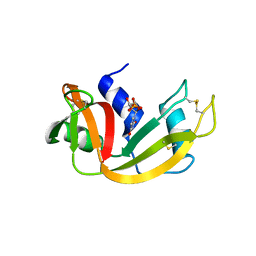 | | Crystal Structure of Bovine Pancreatic Ribonuclease Complexed with Uridine-5'-monophosphate at 1.60 A Resolution | | Descriptor: | Ribonuclease pancreatic, URIDINE-5'-MONOPHOSPHATE | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, Mcpherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic ribonuclease complexed with uridine 5'-monophosphate at 1.60 A resolution.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1A1H
 
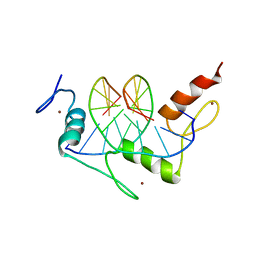 | | QGSR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), QGSR ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
2K7C
 
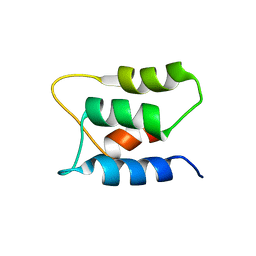 | |
1A5Q
 
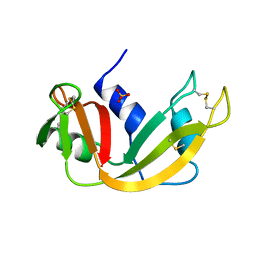 | | P93A VARIANT OF BOVINE PANCREATIC RIBONUCLEASE A | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Pearson, M.A, Karplus, P.A, Dodge, R.W, Laity, J.H, Scheraga, H.A. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two mutants that have implications for the folding of bovine pancreatic ribonuclease A.
Protein Sci., 7, 1998
|
|
7D7V
 
 | |
1AKS
 
 | | CRYSTAL STRUCTURE OF THE FIRST ACTIVE AUTOLYSATE FORM OF THE PORCINE ALPHA TRYPSIN | | Descriptor: | ALPHA TRYPSIN, CALCIUM ION | | Authors: | Johnson, A, Krishnaswamy, S, Sundaram, P.V, Pattabhi, V. | | Deposit date: | 1996-07-24 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure at 1.8 A resolution of an active autolysate form of porcine alpha-trysoin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1A09
 
 | |
1A0U
 
 | | HEMOGLOBIN (VAL BETA1 MET) MUTANT | | Descriptor: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
