8XGT
 
 | |
8XBS
 
 | | C. elegans apo-SID1 structure | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
9LDB
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
9LDT
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
8XC1
 
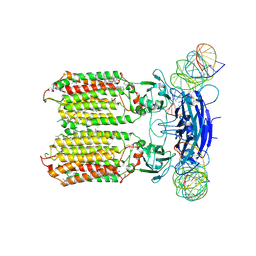 | | C. elegans SID1 in complex with dsRNA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
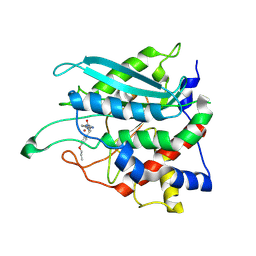
8XGB
 
 | |
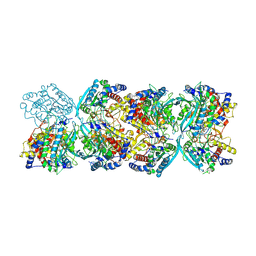
8XGY
 
 | |
7QTT
 
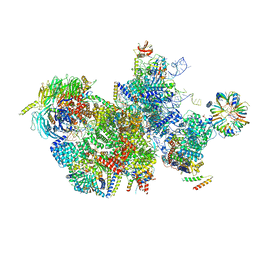 | |
3CZ2
 
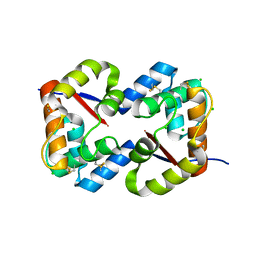 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera at pH 7.0 | | Descriptor: | CHLORIDE ION, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
4U28
 
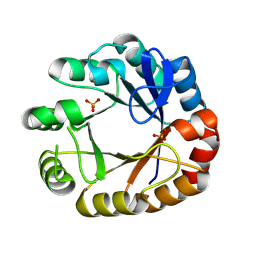 | | Crystal structure of apo Phosphoribosyl isomerase A from Streptomyces sviceus ATCC 29083 | | Descriptor: | PHOSPHATE ION, Phosphoribosyl isomerase A | | Authors: | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4U3B
 
 | |
7R1U
 
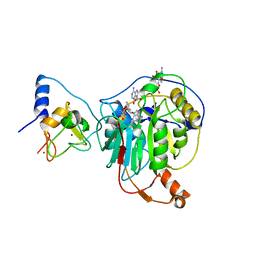 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1T
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R9X
 
 | |
4U3D
 
 | |
4TX9
 
 | | Crystal structure of HisAp from Streptomyces sviceus with degraded ProFAR | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Phosphoribosyl isomerase A, SULFATE ION | | Authors: | Michalska, K, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
7RW2
 
 | |
4U9B
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)NO ligation state | | Descriptor: | GLYCEROL, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4UUK
 
 | | Human dynamin 1 K44A superconstricted polymer stabilized with GTP strand 2 | | Descriptor: | DYNAMIN-1 | | Authors: | Sundborger, A.C, Fang, S, Heymann, J.A, Ray, P, Chappie, J.S, Hinshaw, J.E. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | A Dynamin Mutant Defines a Superconstricted Prefission State.
Cell Rep., 8, 2014
|
|
4W9T
 
 | | Crystal structure of HisAP from Streptomyces sp. Mg1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
7SJ0
 
 | |
7TAA
 
 | | FAMILY 13 ALPHA AMYLASE IN COMPLEX WITH ACARBOSE | | Descriptor: | CALCIUM ION, MODIFIED ACARBOSE HEXASACCHARIDE, TAKA AMYLASE | | Authors: | Davies, G.J, Brzozowski, A.M. | | Deposit date: | 1997-10-06 | | Release date: | 1998-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Aspergillus oryzae alpha-amylase complexed with the inhibitor acarbose at 2.0 A resolution.
Biochemistry, 36, 1997
|
|
7T5F
 
 | |
7SXI
 
 | | Solution Structure of Sds3 Capped Tudor Domain | | Descriptor: | Sin3 histone deacetylase corepressor complex component SDS3 | | Authors: | Marcum, R.D, Radhakrishnan, I. | | Deposit date: | 2021-11-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A capped Tudor domain within a core subunit of the Sin3L/Rpd3L histone deacetylase complex binds to nucleic acid G-quadruplexes.
J.Biol.Chem., 298, 2022
|
|
7ROQ
 
 | | Alternative Structure of Human ABCA1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aller, S.G. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Refined ABCA1 with Domain-swapped Regulatory Domains
Plos One, 2022
|
|
