6DHS
 
 | | Structure of hnRNP H qRRM1,2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein H | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Conformational Dynamics Encoded by the Inter-qRRM linker of hnRNP H.
J. Am. Chem. Soc., 2018
|
|
4CYA
 
 | |
4CHD
 
 | | Crystal structure of the '627' domain of the PB2 subunit of Thogoto virus polymerase | | Descriptor: | POLYMERASE ACIDIC PROTEIN | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
3NFE
 
 | | The crystal structure of hemoglobin I from trematomus newnesi in deoxygenated state | | Descriptor: | Hemoglobin subunit alpha-1, Hemoglobin subunit beta-1/2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vergara, A, Vitagliano, L, Merlino, A, Sica, F, Marino, K, Mazzarella, L. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | An order-disorder transition plays a role in switching off the root effect in fish hemoglobins.
J.Biol.Chem., 285, 2010
|
|
6O64
 
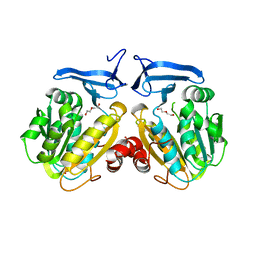 | |
6TA9
 
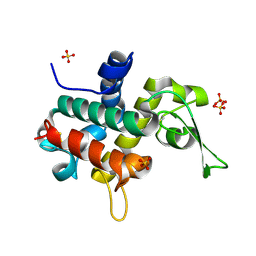 | | Bd0314 DslA wild-type form 1 | | Descriptor: | SLT domain-containing protein, SULFATE ION | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus.
Nat Commun, 11, 2020
|
|
3NN6
 
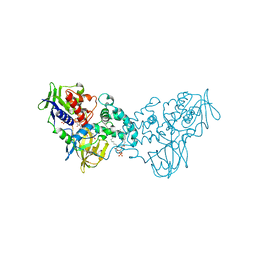 | | Crystal structure of inhibitor-bound in active centre 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, (2R,3S,4S)-5-({[(acetylcarbamoyl)amino]methyl}[(3S,4R)-6-amino-3,4-dimethylhexyl]amino)-2,3,4-trihydroxypentyl [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 5-[(2R)-1-methylpyrrolidin-2-yl]pyridin-2-ol, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-23 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
4DNT
 
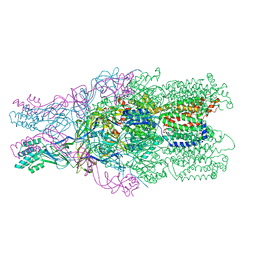 | | Crystal structure of the CusBA heavy-metal efflux complex from Escherichia coli, mutant | | Descriptor: | Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E. | | Deposit date: | 2012-02-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
3N9Z
 
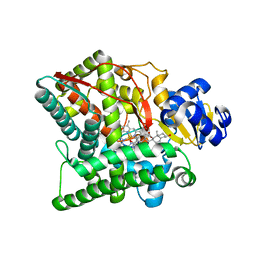 | | Crystal structure of human CYP11A1 in complex with 22-hydroxycholesterol | | Descriptor: | (3alpha,8alpha,22R)-cholest-5-ene-3,22-diol, Adrenodoxin, Cholesterol side-chain cleavage enzyme, ... | | Authors: | Strushkevich, N.V, MacKenzie, F, Tempel, W, Botchkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J.U, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4DFI
 
 | | Crystal structure of cell adhesion molecule nectin-2/CD112 mutant FAMP | | Descriptor: | Poliovirus receptor-related protein 2 | | Authors: | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2012-01-23 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
6DHP
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.04 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6TDX
 
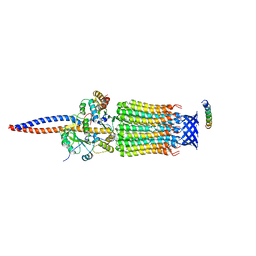 | | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, rotor, rotational state 1 | | Descriptor: | ATP synthase F1 subunit epsilon, ATP synthase F1 subunit gamma, ATP synthase subunit c, ... | | Authors: | Muhleip, A, Amunts, A. | | Deposit date: | 2019-11-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Elife, 8, 2019
|
|
3NCM
 
 | | NEURAL CELL ADHESION MOLECULE, MODULE 2, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (NEURAL CELL ADHESION MOLECULE, LARGE ISOFORM) | | Authors: | Jensen, P.H, Soroka, V, Thomsen, N.K, Berezin, V, Bock, E, Poulsen, F.M. | | Deposit date: | 1998-09-21 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of NCAM modules 1 and 2, basic elements in neural cell adhesion
Nat.Struct.Biol., 6, 1999
|
|
6DRJ
 
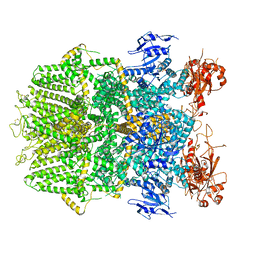 | | Structure of TRPM2 ion channel receptor by single particle electron cryo-microscopy, ADPR/Ca2+ bound state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, Transient receptor potential cation channel, ... | | Authors: | Du, J, Lu, W, Huang, Y, Winkler, P, Sun, W. | | Deposit date: | 2018-06-12 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium.
Nature, 562, 2018
|
|
3NK1
 
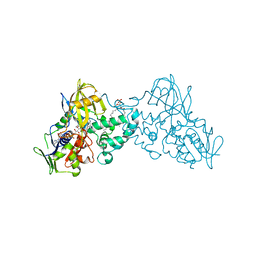 | | Complex of 6-hydroxy-L-nicotine oxidase with serotonin | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-18 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NJQ
 
 | |
4DI3
 
 | |
4DJ7
 
 | | Structure of the hemagglutinin complexed with 3SLN from a highly pathogenic H7N7 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Carney, P.J, Donis, R.O, Stevens, J. | | Deposit date: | 2012-02-01 | | Release date: | 2012-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus.
J.Virol., 86, 2012
|
|
4RDJ
 
 | | Crystal structure of Norovirus Boxer P domain | | Descriptor: | Capsid | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
8JD8
 
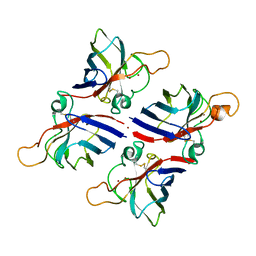 | | Crystal structure of Citrus limon Cu-Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Utami, R.A, Yoshida, H, Retnoningrum, D.S, Ismaya, W.T. | | Deposit date: | 2023-05-12 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Direct relationship between dimeric form and activity in the acidic copper-zinc superoxide dismutase from lemon.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6DF2
 
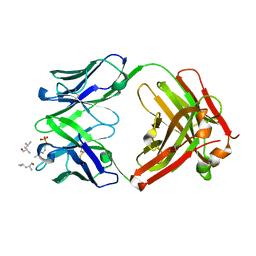 | |
3NTB
 
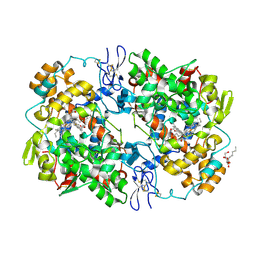 | | Structure of 6-methylthio naproxen analog bound to mCOX-2. | | Descriptor: | (2S)-2-[6-(methylsulfanyl)naphthalen-2-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3NT1
 
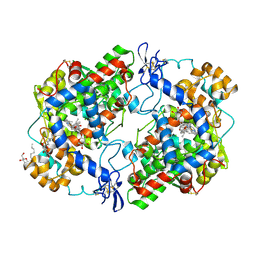 | | High resolution structure of naproxen:COX-2 complex. | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
6DJQ
 
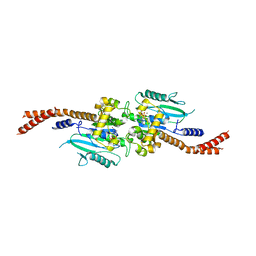 | | Vps1 GTPase-BSE fusion complexed with GDP.AlF4- | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Varlakhanova, N.V, Brady, T.M, Tornabene, B.A, Hosford, C.J, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2018-05-25 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6DRK
 
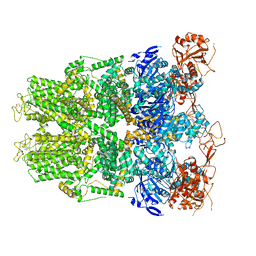 | | Structure of TRPM2 ion channel receptor by single particle electron cryo-microscopy, Apo state | | Descriptor: | Transient receptor potential cation channel, subfamily M, member 2 | | Authors: | Du, J, Lu, W, Huang, Y, Winkler, P, Sun, W. | | Deposit date: | 2018-06-12 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium.
Nature, 562, 2018
|
|
