1J4B
 
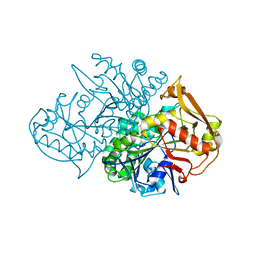 | | Recombinant Mouse-Muscle Adenylosuccinate Synthetase | | Descriptor: | adenylosuccinate synthetase | | Authors: | Iancu, C.V, Borza, T, Choe, J.Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-09-07 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recombinant mouse muscle adenylosuccinate synthetase: overexpression, kinetics, and crystal structure.
J.Biol.Chem., 276, 2001
|
|
1H4L
 
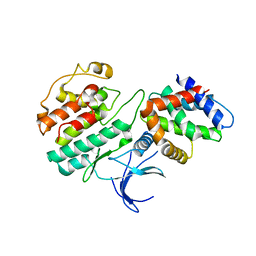 | | Structure and regulation of the CDK5-p25(nck5a) complex | | Descriptor: | CELL DIVISION PROTEIN KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR | | Authors: | Tarricone, C, Dhavan, R, Peng, J, Areces, L.B, Tsai, L.-H, Musacchio, A. | | Deposit date: | 2001-05-11 | | Release date: | 2002-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Regulation of the Cdk5-P25(Nck5A) Complex
Mol.Cell, 8, 2001
|
|
1ISI
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with ethenoNAD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, [[(2R,3S,4R,5R)-3,4-dihydroxy-5-(9H-imidazo[2,1-f]purin-6-ium-3-yl)oxolan-2-yl]methoxy-oxidanidyl-phosphoryl] [(2R,3S,4R,5R)-3,4-dihydroxy-5-oxidanidyl-oxolan-2-yl]methyl phosphate, ... | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1M1X
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHA VBETA3 BOUND TO MN2+ | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.-P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.
Science, 296, 2002
|
|
1H4R
 
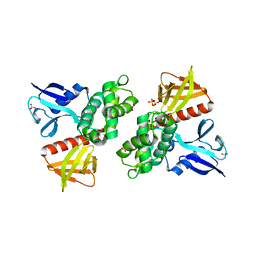 | | Crystal Structure of the FERM domain of Merlin, the Neurofibromatosis 2 Tumor Suppressor Protein. | | Descriptor: | MERLIN, SULFATE ION | | Authors: | Cooper, D.R, Kang, B.S, Sheffield, P, Devedjiev, Y, Derewenda, Z.S. | | Deposit date: | 2001-05-14 | | Release date: | 2002-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Ferm Domain of Merlin, the Neurofibromatosis Type 2 Gene Product.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1MEZ
 
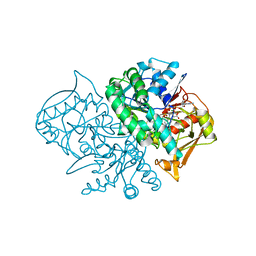 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with SAMP, GDP, SO4(2-), and Mg(2+) | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1MF0
 
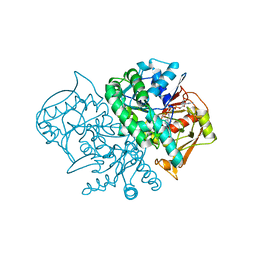 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP, GDP, HPO4(2-), and Mg(2+) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1M45
 
 | |
7MWL
 
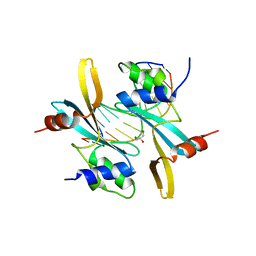 | | The TAM domain of BAZ2A in complex with a 12mer mCG DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), GLYCEROL | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The TAM domain of BAZ2A in complex with a 12mer mCG DNA
To Be Published
|
|
7MGJ
 
 | |
7MGK
 
 | |
7MWH
 
 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the BAZ2B TAM domain.
Heliyon, 8, 2022
|
|
7OZH
 
 | |
7OZG
 
 | |
7PQH
 
 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7LZB
 
 | | Crystal Structure of SETD2 bound to Compound 2 | | Descriptor: | BETA-MERCAPTOETHANOL, Histone-lysine N-methyltransferase SETD2, N-[(1r,4r)-4-(beta-alanylamino)cyclohexyl]-7-methyl-1H-indole-2-carboxamide, ... | | Authors: | Farrow, N.A, Boriack-Sjodin, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the Histone Methyltransferase SETD2 Suitable for Preclinical Studies.
Acs Med.Chem.Lett., 12, 2021
|
|
7LZD
 
 | | Crystal Structure of SETD2 bound to Compound 35 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETD2, ... | | Authors: | Farrow, N.A, Boriack-Sjodin, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the Histone Methyltransferase SETD2 Suitable for Preclinical Studies.
Acs Med.Chem.Lett., 12, 2021
|
|
7LZF
 
 | | Crystal Structure of SETD2 bound to Compound 57 | | Descriptor: | 1,2-ETHANEDIOL, 4-fluoro-N-[(1R,3S)-3-{(3S)-3-[(methanesulfonyl)(methyl)amino]pyrrolidin-1-yl}cyclohexyl]-7-methyl-1H-indole-2-carboxamide, Histone-lysine N-methyltransferase SETD2, ... | | Authors: | Farrow, N.A, Boriack-Sjodin, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the Histone Methyltransferase SETD2 Suitable for Preclinical Studies.
Acs Med.Chem.Lett., 12, 2021
|
|
3UE2
 
 | |
3UWT
 
 | |
5OOW
 
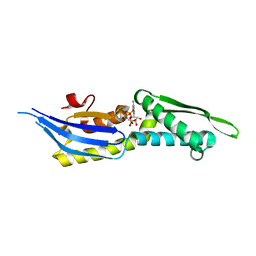 | | Crystal structure of lobe II from the nucleotide binding domain of DnaK in complex with AMPPCP | | Descriptor: | Chaperone protein DnaK, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Jakob, R.P, Bauer, D, Meinhold, S, Stigler, J, Merkel, U, Maier, T, Rief, M, Zoldak, G. | | Deposit date: | 2017-08-09 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A folding nucleus and minimal ATP binding domain of Hsp70 identified by single-molecule force spectroscopy.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4OWW
 
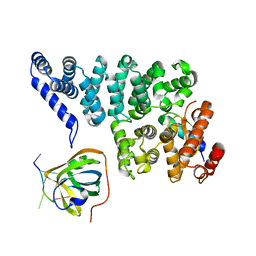 | | Structural basis of SOSS1 in complex with a 35nt ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), Integrator complex subunit 3, SOSS complex subunit B1, ... | | Authors: | Ren, W, Sun, Q, Tang, X, Song, H. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA.
Cell Rep, 6, 2014
|
|
3JUA
 
 | |
7UX2
 
 | |
6D94
 
 | | Crystal structure of PPAR gamma in complex with Mediator of RNA polymerase II transcription subunit 1 | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | Deposit date: | 2018-04-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural mechanism of nuclear receptor biased agonism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
