6SIU
 
 | | Crystal structure of IbpAFic2 covalently tethered to Cdc42 | | Descriptor: | Cell division control protein 42 homolog, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gulen, B, Roselin, M, Albers, M, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2019-08-12 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Identification of targets of AMPylating Fic enzymes by co-substrate-mediated covalent capture.
Nat.Chem., 12, 2020
|
|
6EEI
 
 | | Crystal structure of Arabidopsis thaliana phenylacetaldehyde synthase in complex with L-phenylalanine | | Descriptor: | PHENYLALANINE, SULFATE ION, Tyrosine decarboxylase 1 | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (1.99001348 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEW
 
 | | Crystal structure of Catharanthus roseus tryptophan decarboxylase in complex with L-tryptophan | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CALCIUM ION, TRYPTOPHAN | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.05002069 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UR0
 
 | | Crystal structure of ChoE D285N mutant acyl-enzyme | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ChoE, GLYCEROL | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the putative bacterial acetylcholinesterase ChoE and its substrate inhibition mechanism.
J.Biol.Chem., 295, 2020
|
|
3NR8
 
 | | Crystal structure of human SHIP2 | | Descriptor: | CHLORIDE ION, Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases
Structure, 22, 2014
|
|
1ROC
 
 | | Crystal structure of the histone deposition protein Asf1 | | Descriptor: | Anti-silencing protein 1, BROMIDE ION | | Authors: | Daganzo, S.M, Erzberger, J.P, Lam, W.M, Skordalakes, E, Zhang, R, Franco, A.A, Brill, S.J, Adams, P.D, Berger, J.M, Kaufman, P.D. | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the conserved core of histone deposition protein Asf1.
Curr.Biol., 13, 2003
|
|
1RP5
 
 | | PBP2x from Streptococcus pneumoniae strain 5259 with reduced susceptibility to beta-lactam antibiotics | | Descriptor: | SULFATE ION, penicillin-binding protein 2x | | Authors: | Pernot, L, Chesnel, L, Legouellec, A, Croize, J, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics.
J.Biol.Chem., 279, 2004
|
|
6JJA
 
 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP | | Descriptor: | CALCIUM ION, Nucleocapsid protein CP17 | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP
To Be Published
|
|
7BCM
 
 | | The DDR1 Kinase Domain Bound To SR302 | | Descriptor: | Epithelial discoidin domain-containing receptor 1, ~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{S})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Roehm, S, Joerger, A, Knapp, S. | | Deposit date: | 2020-12-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
7BNV
 
 | | Crystal Structure of the SARS-CoV-2 Receptor Binding Domain in Complex with Antibody ION-300 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain, Light Chain, ... | | Authors: | Hall, G, Cowan, R, Carr, M. | | Deposit date: | 2021-01-22 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cross-Reactive SARS-CoV-2 Neutralizing Antibodies From Deep Mining of Early Patient Responses.
Front Immunol, 12, 2021
|
|
1KNU
 
 | | LIGAND BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA IN COMPLEX WITH A SYNTHETIC AGONIST | | Descriptor: | (S)-3-(4-(2-CARBAZOL-9-YL-ETHOXY)-PHENYL)-2-ETHOXY-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA | | Authors: | Svensson, L.A, Mortensen, S.B, Fleckner, J, Woeldike, H.F. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclic-alpha-alkyloxyphenylpropionic acids: dual PPARalpha/gamma agonists with hypolipidemic and antidiabetic activity
J.MED.CHEM., 45, 2002
|
|
8DP6
 
 | |
8DP7
 
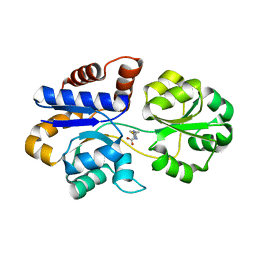 | |
6ZUL
 
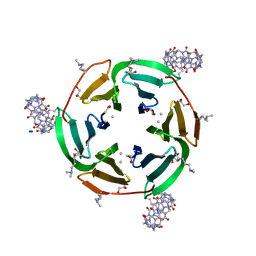 | | Crystal structure of dimethylated RSL in complex with cucurbit[7]uril and zinc | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SODIUM ION, ... | | Authors: | Guagnini, F, Engilberge, S, Flood, R.J, Ramberg, K.O, Crowley, P.B. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Metal-Mediated Protein-Cucurbituril Crystalline Architectures
Cryst.Growth Des., 2020
|
|
6ZUM
 
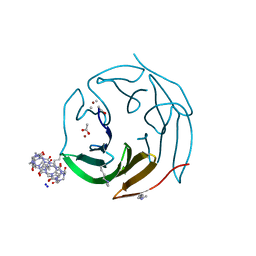 | | Crystal structure of dimethylated RSL-N23H (RSL-B3) in complex with cucurbit[7]uril and zinc | | Descriptor: | ACETATE ION, Fucose-binding lectin protein, SODIUM ION, ... | | Authors: | Guagnini, F, Engilberge, S, Flood, R.J, Ramberg, K.O, Crowley, P.B. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Metal-Mediated Protein-Cucurbituril Crystalline Architectures
Cryst.Growth Des., 2020
|
|
4JL6
 
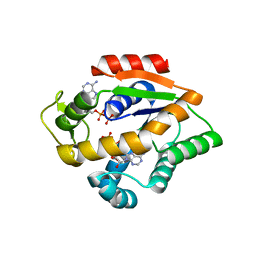 | |
1S01
 
 | |
4JL5
 
 | |
7TDB
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine bisphosphonate, manganese ions | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, [2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethoxy-oxidanyl-phosphoryl]methylphosphonic acid, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery.
Acs Chem.Biol., 17, 2022
|
|
7TD7
 
 | |
7TDC
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine bisphosphonate, calcium ions | | Descriptor: | CALCIUM ION, MAGNESIUM ION, [2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethoxy-oxidanyl-phosphoryl]methylphosphonic acid, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery.
Acs Chem.Biol., 17, 2022
|
|
7TDA
 
 | |
8RFG
 
 | | CgsiGP3 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, ... | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
8RF0
 
 | | WT-CGS sample in nanodisc | | Descriptor: | Cyclic beta-(1,2)-glucan synthase NdvB, URIDINE-5'-DIPHOSPHATE-GLUCOSE, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, ... | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
