5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
2AN9
 
 | | Crystal Structure Of Oligomeric E.coli Guanylate Kinase In Complex With GDP | | Descriptor: | GUANOSINE, GUANOSINE-5'-DIPHOSPHATE, Guanylate kinase, ... | | Authors: | Hible, G, Renault, L, Schaeffer, F, Christova, P, Radulescu, A.Z, Evrin, C, Gilles, A.M, Cherfils, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase
J.Mol.Biol., 352, 2005
|
|
2H8I
 
 | |
5E79
 
 | | Macromolecular diffractive imaging using imperfect crystals | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthur, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffer, A, Dorner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
2D1L
 
 | | Structure of F-actin binding domain IMD of MIM (Missing In Metastasis) | | Descriptor: | Metastasis suppressor protein 1 | | Authors: | Lee, S.H, Kerff, F, Chereau, D, Ferron, F, Dominguez, R. | | Deposit date: | 2005-08-27 | | Release date: | 2006-09-12 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the actin-binding function of missing-in-metastasis
Structure, 15, 2007
|
|
2D1K
 
 | | Ternary complex of the WH2 domain of mim with actin-dnase I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chereau, D, Kerff, F, Dominguez, R. | | Deposit date: | 2005-08-26 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the actin-binding function of missing-in-metastasis
Structure, 15, 2007
|
|
2WKE
 
 | | Crystal structure of the Actinomadura R39 DD-peptidase inhibited by 6- beta-iodopenicillanate. | | Descriptor: | (3S)-2,2-dimethyl-3,4-dihydro-2H-1,4-thiazine-3,6-dicarboxylic acid, COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2009-06-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of the Inhibition of Class a Beta-Lactamases and Penicillin-Binding Proteins by 6-Beta-Iodopenicillanate.
J.Am.Chem.Soc., 131, 2009
|
|
1R8S
 
 | | ARF1[DELTA1-17]-GDP IN COMPLEX WITH A SEC7 DOMAIN CARRYING THE MUTATION OF THE CATALYTIC GLUTAMATE TO LYSINE | | Descriptor: | ADP-ribosylation factor 1, Arno, BETA-MERCAPTOETHANOL, ... | | Authors: | Renault, L, Guibert, B, Cherfils, J. | | Deposit date: | 2003-10-28 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor
Nature, 426, 2003
|
|
1S9D
 
 | | ARF1[DELTA 1-17]-GDP-MG IN COMPLEX WITH BREFELDIN A AND A SEC7 DOMAIN | | Descriptor: | 1,6,7,8,9,11A,12,13,14,14A-DECAHYDRO-1,13-DIHYDROXY-6-METHYL-4H-CYCLOPENT[F]OXACYCLOTRIDECIN-4-ONE, ADP-Ribosylation Factor 1, Arno, ... | | Authors: | Renault, L, Guibert, B, Cherfils, J. | | Deposit date: | 2004-02-04 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Snapshots of the Mechanism and Inhibition of a Guanine Nucleotide Exchange Factor
Nature, 426, 2003
|
|
5DJB
 
 | |
1R8M
 
 | |
1R8Q
 
 | | FULL-LENGTH ARF1-GDP-MG IN COMPLEX WITH BREFELDIN A AND A SEC7 DOMAIN | | Descriptor: | 1,6,7,8,9,11A,12,13,14,14A-DECAHYDRO-1,13-DIHYDROXY-6-METHYL-4H-CYCLOPENT[F]OXACYCLOTRIDECIN-4-ONE, ADP-ribosylation factor 1, Arno, ... | | Authors: | Renault, L, Guibert, B, Cherfils, J. | | Deposit date: | 2003-10-28 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor
Nature, 426, 2003
|
|
2WK0
 
 | | Crystal structure of the class A beta-lactamase BS3 inhibited by 6- beta-iodopenicillanate. | | Descriptor: | (3S)-2,2-dimethyl-3,4-dihydro-2H-1,4-thiazine-3,6-dicarboxylic acid, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Sauvage, E, Zervosen, A, Dive, G, Herman, R, Kerff, F, Amoroso, A, Fonze, E, Pratt, R.F, Luxen, A, Charlier, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of the Inhibition of Class a Beta-Lactamases and Penicillin-Binding Proteins by 6-Beta-Iodopenicillanate.
J.Am.Chem.Soc., 131, 2009
|
|
2LMS
 
 | | A single GalNAc residue on Threonine-106 modifies the dynamics and the structure of Interferon alpha-2a around the glycosylation site | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Interferon alpha-2 | | Authors: | Ghasriani, H, Belcourt, P.J.F, Sauve, S, Hodgson, D.J, Gingras, G, Brochu, D, Gilbert, M, Aubin, Y. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A single N-acetylgalactosamine residue at threonine 106 modifies the dynamics and structure of interferon alpha2a around the glycosylation site.
J.Biol.Chem., 288, 2013
|
|
2WKX
 
 | | Crystal structure of the native E. coli zinc amidase AmiD | | Descriptor: | CHLORIDE ION, GLYCEROL, N-ACETYLMURAMOYL-L-ALANINE AMIDASE AMID, ... | | Authors: | Petrella, S, Kerff, F, Herman, R, Genereux, C, Pennartz, A, Sauvage, E, Joris, B, Charlier, P. | | Deposit date: | 2009-06-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-L-Alanine Amidase Amid from Escherichia Coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
1MY6
 
 | | The 1.6 A Structure of Fe-Superoxide Dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus : Correlation of EPR and Structural Characteristics | | Descriptor: | FE (III) ION, Iron (III) Superoxide Dismutase | | Authors: | Yoshida, S.M, Cascio, D, Sawaya, M.R, Yeates, T.O, Kerfeld, C.A. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A resolution structure of Fe-superoxide dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus.
J.BIOL.INORG.CHEM., 8, 2003
|
|
2M0V
 
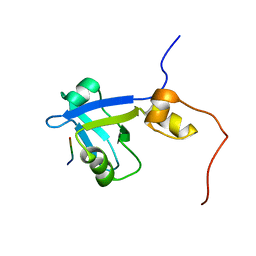 | | Complex structure of C-terminal CFTR peptide and extended PDZ2 domain from NHERF1 | | Descriptor: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | Deposit date: | 2012-11-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
2M0T
 
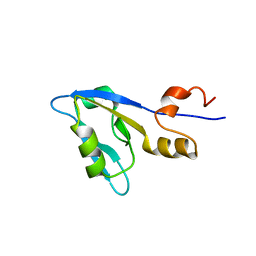 | |
4KA2
 
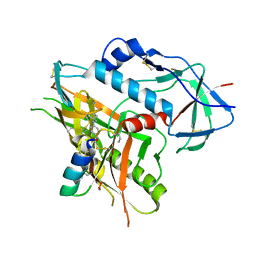 | |
2M0U
 
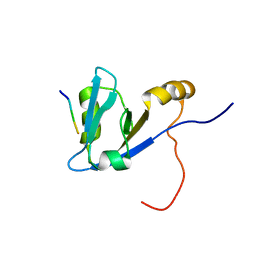 | | Complex structure of C-terminal CFTR peptide and extended PDZ1 domain from NHERF1 | | Descriptor: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | Deposit date: | 2012-11-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
2KT8
 
 | | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B | | Descriptor: | Probable surface protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B
To be Published
|
|
2KRS
 
 | | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A. | | Descriptor: | Probable enterotoxin | | Authors: | Ramelot, T.A, Cort, J.R, Maglaqui, M, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A.
To be Published
|
|
2JNK
 
 | |
2NVP
 
 | | X-Ray Crystal Structure of Protein CPF_0428 from Clostridium perfringens. Northeast Structural Genomics Consortium Target CpR63. | | Descriptor: | Hypothetical protein | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Cunningham, K, Ma, L.-C, Fang, Y, Baran, M.C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-13 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Hypothetical Protein CPF_0428 from Clostridium perfringens, Northeast Structural Genomics Target CpR63.
TO BE PUBLISHED
|
|
2JOV
 
 | | NMR Structure of Clostridium Perfringens Protein CPE0013. Northeast Structural Genomics Target CpR31. | | Descriptor: | Hypothetical protein CPE0013 | | Authors: | Ding, K, Ramelot, T.A, Anklin, C.G, Wang, H, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-04 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Clostridium perfringens protein CPE0013.
To be Published
|
|
