400D
 
 | |
2SPZ
 
 | | STAPHYLOCOCCAL PROTEIN A, Z-DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN A | | Authors: | Montelione, G.T, Tashiro, M, Tejero, R, Lyons, B.A. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution NMR structure of the Z domain of staphylococcal protein A.
J.Mol.Biol., 272, 1997
|
|
1SCZ
 
 | | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase | | Descriptor: | Dihydrolipoamide Succinyltransferase | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Tsao, J, Johnson, D, Huang, W.-Y, Pruett, P, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, Z, Lu, S, DeLucas, L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase
To be Published
|
|
1AN2
 
 | | RECOGNITION BY MAX OF ITS COGNATE DNA THROUGH A DIMERIC B/HLH/Z DOMAIN | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*GP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*C P*TP*AP*CP*AP*C)- 3'), PROTEIN (TRANSCRIPTION FACTOR MAX (TF MAX)) | | Authors: | Ferre-D'Amare, A.R, Prendergast, G.C, Ziff, E.B, Burley, S.K. | | Deposit date: | 1996-09-06 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain.
Nature, 363, 1993
|
|
3A9F
 
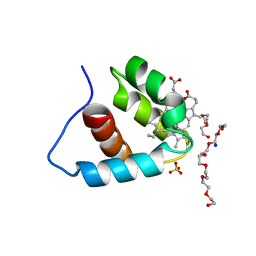 | | Crystal structure of the C-terminal domain of cytochrome cz from Chlorobium tepidum | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Cytochrome c, HEME C, ... | | Authors: | Hirano, Y, Higuchi, M, Azai, C, Oh-oka, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2009-10-25 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the electron carrier domain of the reaction center cytochrome c(z) subunit from green photosynthetic bacterium Chlorobium tepidum
J.Mol.Biol., 397, 2010
|
|
2ELG
 
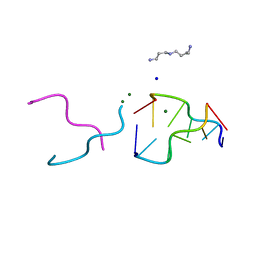 | | The rare crystallographic structure of d(CGCGCG)2: The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)2 at 10 degree celsius | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Tozuka, Y, Zhou, D.Y, Ishida, T, Nakatani, K. | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The rare crystallographic structure of d(CGCGCG)(2): The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)(2) at 10 degrees C
Biochem.Biophys.Res.Commun., 358, 2007
|
|
4V41
 
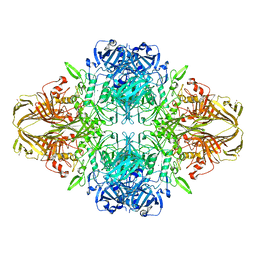 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
5WSN
 
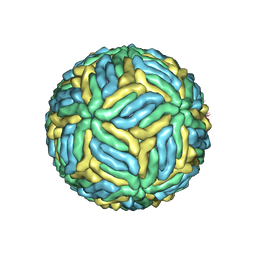 | | Structure of Japanese encephalitis virus | | Descriptor: | E protein, M protein | | Authors: | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
6SCZ
 
 | | Mycobacterium tuberculosis alanine racemase inhibited by DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | de Chiara, C, Purkiss, A, Prosser, G, Homsak, M, de Carvalho, L.P.S. | | Deposit date: | 2019-07-26 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | D-Cycloserine destruction by alanine racemase and the limit of irreversible inhibition.
Nat.Chem.Biol., 16, 2020
|
|
3VD9
 
 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
3VD3
 
 | | E. coli (lacZ) beta-galactosidase (N460D) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
3VD7
 
 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with galactotetrazole | | Descriptor: | (5R, 6S, 7S, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
3VDB
 
 | | E. coli (lacZ) beta-galactosidase (N460T) in complex with galactonolactone | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
3VDC
 
 | | E. coli (lacZ) beta-galactosidase (N460T) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
3VDA
 
 | | E. coli (lacZ) beta-galactosidase (N460T) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
4N99
 
 | | E. coli sliding clamp in complex with 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid | | Descriptor: | 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N9A
 
 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N97
 
 | | E. coli sliding clamp in complex with 5-nitroindole | | Descriptor: | 5-nitro-1H-indole, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
3N3G
 
 | | 4-(3-Trifluoromethylphenyl)-pyrimidine-2-carbonitrile as cathepsin S inhibitors: N3, not N1 is critically important | | Descriptor: | (E)-1-(6-{4-[3-(4-methylpiperazin-1-yl)propoxy]-3-(trifluoromethyl)phenyl}pyridin-2-yl)methanimine, 6-{4-[3-(4-methylpiperazin-1-yl)propoxy]-3-(trifluoromethyl)phenyl}pyridine-2-carbonitrile, Cathepsin S, ... | | Authors: | Fradera, X, Uitdehaag, J.C.M, van Zeeland, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-07-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 4-(3-Trifluoromethylphenyl)-pyrimidine-2-carbonitrile as cathepsin S inhibitors: N3, not N1 is critically important.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8UZG
 
 | | E. coli 70S ribosome with unmodified e*/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-11-15 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
8V03
 
 | | E. coli 70S ribosome with unmodified P/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-11-16 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
8UTJ
 
 | | E. coli 70S ribosome with unmodified lys-tRNAPro(GGG) bound to slippery P-site CCC-C codon in the 0 frame | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-10-31 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
8UXB
 
 | | E. coli 70S ribosome with unmodified P/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-11-09 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
8URM
 
 | | E. coli 70S ribosome with unmodified tRNAPro(GGG) bound to slippery P-site CCC-C codon and tRNAVal(UAC) in the A site | | Descriptor: | 13S ribosomal RNA, 16S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-10-26 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
8UX8
 
 | | E. coli 70S ribosome with unmodified Lys-tRNAPro(GGG) bound to slippery P-site CCC-C codon in the +1 mRNA reading frame | | Descriptor: | 16S ribosomal RNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-11-09 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
