6B82
 
 | | Zebra Fish CYP-450 17A1 Mutant Abiraterone Complex | | Descriptor: | ACETATE ION, Abiraterone, CHLORIDE ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Inherent steroid 17 alpha ,20-lyase activity in defunct cytochrome P450 17A enzymes.
J. Biol. Chem., 293, 2018
|
|
7WDD
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87K in complex with N-imidazolyl-hexanoyl-L-phenylalanine, styrene and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
7WDG
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7Z38
 
 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7Z37
 
 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
5IKV
 
 | | The Structure of Flufenamic Acid Bound to Human Cyclooxygenase-2 | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Orlando, B.J, Malkowski, M.G. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Substrate-selective Inhibition of Cyclooxygeanse-2 by Fenamic Acid Derivatives Is Dependent on Peroxide Tone.
J.Biol.Chem., 291, 2016
|
|
5IKR
 
 | | The Structure of Mefenamic Acid Bound to Human Cyclooxygenase-2 | | Descriptor: | 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Orlando, B.J, Malkowski, M.G. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Substrate-selective Inhibition of Cyclooxygeanse-2 by Fenamic Acid Derivatives Is Dependent on Peroxide Tone.
J.Biol.Chem., 291, 2016
|
|
5IFS
 
 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | Deposit date: | 2016-02-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
5ISM
 
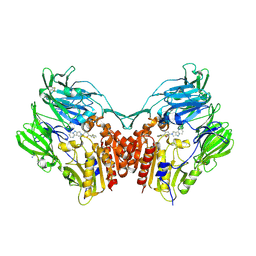 | | Human DPP4 in complex with a novel 5,5,6-tricyclic pyrrolidine inhibitor | | Descriptor: | (2R,3S,5R)-2-(2,5-difluorophenyl)-5-(7H-pyrrolo[3',4':3,4]pyrazolo[1,5-a]pyrimidin-8(9H)-yl)oxan-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-03-15 | | Release date: | 2016-05-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery of novel 5,6,5- and 5,5,6-tricyclic pyrrolidines as potent and selective DPP-4 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5IL1
 
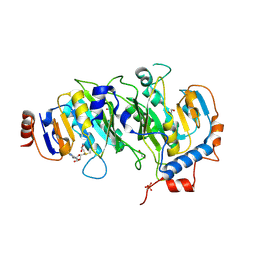 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL0
 
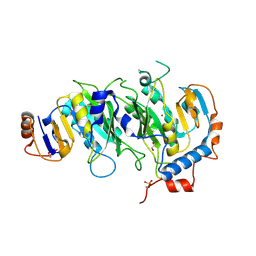 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5SYC
 
 | |
5SZG
 
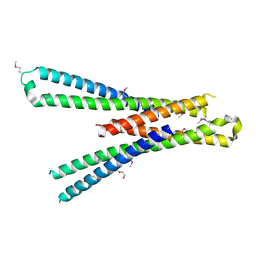 | | Structure of the bMERB domain of Mical-3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein-methionine sulfoxide oxidase MICAL3 | | Authors: | Rai, A, Oprisko, A, Campos, J, Fu, Y, Friese, T, Itzen, A, Goody, R.S, Gazdag, E.M, Mueller, M.P. | | Deposit date: | 2016-08-14 | | Release date: | 2016-08-24 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | bMERB domains are bivalent Rab8 family effectors evolved by gene duplication.
Elife, 5, 2016
|
|
5T8O
 
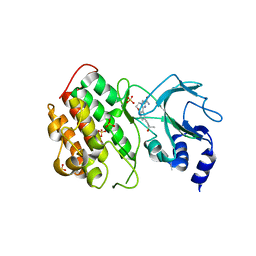 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to Imidazobenzoxepin Compound 3 | | Descriptor: | 10-(3-methyl-3-oxidanyl-but-1-ynyl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Smith, M.A, McEwan, P, Hymowitz, S.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
5T8P
 
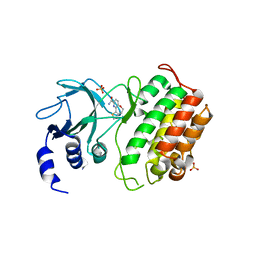 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to benzoxepin compound 2 | | Descriptor: | 6,7-dihydrothieno[4,5]oxepino[1,2-~{c}]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Smith, M.A, McEwan, P.A. | | Deposit date: | 2016-09-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
5J1W
 
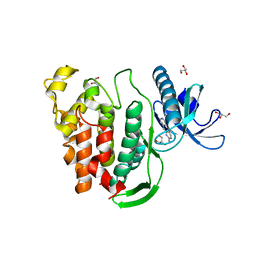 | | Crystal structure of human CLK1 in complex with pyrido[3,4-g]quinazoline derivative ZW31 (compound 14) | | Descriptor: | Dual specificity protein kinase CLK1, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Esvan, Y.J, Zeinyeh, W, Boibessot, T, Nauton, L, Thery, V, Loaec, N, Meijer, L, Giraud, F, Moreau, P, Anizon, F, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of pyrido[3,4-g]quinazoline derivatives as CMGC family protein kinase inhibitors: Design, synthesis, inhibitory potency and X-ray co-crystal structure.
Eur.J.Med.Chem., 118, 2016
|
|
5J3J
 
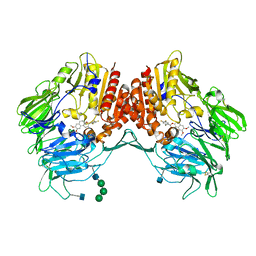 | | Crystal Structure of human DPP-IV in complex with HL1 | | Descriptor: | (2~{S},3~{R})-8,9-dimethoxy-3-[2,4,5-tris(fluoranyl)phenyl]-2,3-dihydro-1~{H}-benzo[f]chromen-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Wu, F, Li, H, Zhao, Z, Zhu, L, Xu, H, Li, S. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of human DPP-IV in complex with HL1
To Be Published
|
|
5U6Y
 
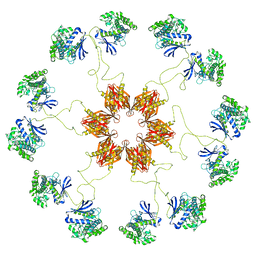 | | Pseudo-atomic model of the CaMKIIa holoenzyme. | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Myers, J, Reichow, S.L. | | Deposit date: | 2016-12-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | The CaMKII holoenzyme structure in activation-competent conformations.
Nat Commun, 8, 2017
|
|
5JWG
 
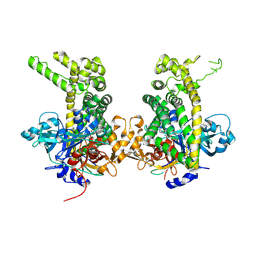 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with dipeptide Arg-Asp | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, ASPARTIC ACID, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K4L
 
 | |
5JXK
 
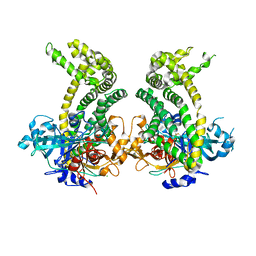 | | Crystal structure of Porphyromonas endodontalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5TV5
 
 | | BioW from Aquifex aeoulicus | | Descriptor: | 6-carboxyhexanoate--CoA ligase | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5UGS
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT501 | | Descriptor: | 5-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]-2-(2-methylphenoxy)phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5JY0
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with substrate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Asp/Glu-specific dipeptidyl-peptidase, LEU-ASP-VAL | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5UDA
 
 | |
