1OR5
 
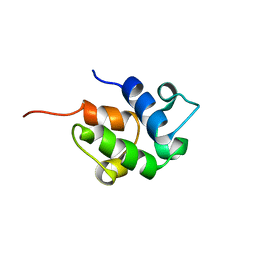 | | SOLUTION STRUCTURE OF THE HOLO-FORM OF THE FRENOLICIN ACYL CARRIER PROTEIN, MINIMIZED MEAN STRUCTURE | | Descriptor: | acyl carrier protein | | Authors: | Li, Q, Khosla, C, Puglisi, J, Liu, C.W. | | Deposit date: | 2003-03-11 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Holo Form of the Frenolicin Acyl Carrier Protein
Biochemistry, 42, 2003
|
|
4I45
 
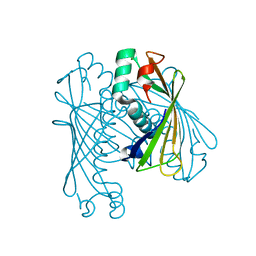 | | Crystal Structure of Orf6 protein from Photobacterium profundum, Mg2+-bound form | | Descriptor: | MAGNESIUM ION, ORF6 thioesterase | | Authors: | Rodriguez-Guilbe, M.M, Baerga-Ortiz, A, Schreiter, E.R. | | Deposit date: | 2012-11-27 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, Activity, and Substrate Selectivity of the Orf6 Thioesterase from Photobacterium profundum.
J.Biol.Chem., 288, 2013
|
|
1CX2
 
 | |
6ZTC
 
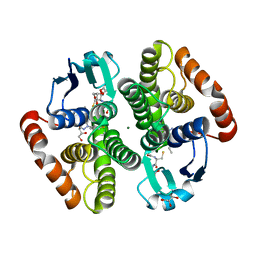 | | CRYSTAL STRUCTURE OF PROSTAGLANDIN D2 SYNTHASE IN COMPLEX WITH FRAGMENT 1A AT 1.84A RESOLUTION. | | Descriptor: | 1-[1-(3-fluorophenyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]propan-1-one, GLUTATHIONE, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
3G0Y
 
 | | Structure of E. coli FabF(C163Q) in complex with dihydroplatensimycin | | Descriptor: | 3-({3-[(1S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxodecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3G11
 
 | | Structure of E. coli FabF(C163Q) in complex with dihydrophenyl platensimycin | | Descriptor: | 3-({3-[(1S,4S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxo-4-phenyldecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3LN0
 
 | | Structure of compound 5c-S bound at the active site of COX-2 | | Descriptor: | (2S)-6,8-dichloro-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kiefer, J.R, Kurumbail, R.G, Stallings, W.C, Pawlitz, J.L. | | Deposit date: | 2010-02-01 | | Release date: | 2010-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6B2S
 
 | | Crystal structure of Xanthomonas campestris OleA H285N | | Descriptor: | 3-oxoacyl-[ACP] synthase III, GLYCEROL, PHOSPHATE ION | | Authors: | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
6B2R
 
 | | Crystal structure of Xanthomonas campestris OleA H285A | | Descriptor: | 3-oxoacyl-[ACP] synthase III, GLYCEROL | | Authors: | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
4RZQ
 
 | | Structural Analysis of Substrate, Reaction Intermediate and Product Binding in Haemophilus influenzae Biotin Carboxylase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, methyl (3aS,4S,6aR)-4-(5-methoxy-5-oxopentyl)-2-oxohexahydro-1H-thieno[3,4-d]imidazole-1-carboxylate | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R, Waldrop, G.L. | | Deposit date: | 2014-12-23 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
8QM1
 
 | | wild type Pa.FabF in complex cerulenin | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
6UUZ
 
 | |
2X3E
 
 | | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III, FabH from Pseudomonas aeruginosa PAO1 | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 3 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2NZ2
 
 | | Crystal structure of human argininosuccinate synthase in complex with aspartate and citrulline | | Descriptor: | ASPARTIC ACID, Argininosuccinate synthase, CITRULLINE, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human argininosuccinate synthetase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7TWA
 
 | | Crystal structure of apo BesC from Streptomyces cattleya | | Descriptor: | 1,3-BUTANEDIOL, 4-chloro-allylglycine synthase, ACETATE ION, ... | | Authors: | Neugebauer, M.E, McBride, M.J, Boal, A.K, Chang, M.C.Y. | | Deposit date: | 2022-02-07 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate-Triggered mu-Peroxodiiron(III) Intermediate in the 4-Chloro-l-Lysine-Fragmenting Heme-Oxygenase-like Diiron Oxidase (HDO) BesC: Substrate Dissociation from, and C4 Targeting by, the Intermediate.
Biochemistry, 61, 2022
|
|
5Y08
 
 | |
6CHJ
 
 | |
3NTG
 
 | | Crystal structure of COX-2 with selective compound 23d-(R) | | Descriptor: | (2R)-6,8-dichloro-7-(2-methylpropoxy)-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.L, Limburg, D, Graneto, M.J, Carter, J.C, Talley, J.J, Kiefer, J.R. | | Deposit date: | 2010-07-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2W6N
 
 | |
2UUV
 
 | | alkyldihydroxyacetonephosphate synthase in P1 | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, FLAVIN-ADENINE DINUCLEOTIDE, HEXADECAN-1-OL | | Authors: | Razeto, A, Mattiroli, F, Carpanelli, E, Aliverti, A, Pandini, V, Coda, A, Mattevi, A. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Crucial Step in Ether Phospholipid Biosynthesis: Structural Basis of a Noncanonical Reaction Associated with a Peroxisomal Disorder.
Structure, 15, 2007
|
|
2W70
 
 | |
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | Descriptor: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
 | |
2V59
 
 | |
2W6Z
 
 | |
