8ZYJ
 
 | | Cryo-EM structure of human testis-specific Na+,K+-ATPase alpha4 in ouabain-bound form | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Abe, K, Blanco, G. | | Deposit date: | 2024-06-18 | | Release date: | 2024-12-04 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Molecular Structure of the Na + ,K + -ATPase alpha 4 beta 1 Isoform in Its Ouabain-Bound Conformation.
Int J Mol Sci, 25, 2024
|
|
5UEJ
 
 | |
5UGD
 
 | | Protease Inhibitor | | Descriptor: | Nalpha-[trans-4-(aminomethyl)cyclohexane-1-carbonyl]-N-octyl-O-[(pyridin-4-yl)methyl]-L-tyrosinamide, Plasminogen | | Authors: | Law, R.H.P, Wu, G, Whisstock, J.C. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | X-ray crystal structure of plasmin with tranexamic acid-derived active site inhibitors.
Blood Adv, 1, 2017
|
|
6KVK
 
 | | Crystal structure of UDP-Sm-SrUGT76G1 | | Descriptor: | Steviolmonoside, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
7CCU
 
 | |
8S1T
 
 | | BzdNO-cyclohexa-1,5-diene-1-carboxy-CoA complex | | Descriptor: | 1,5 Dienoyl-CoA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BzdN, ... | | Authors: | Ermler, U, Boll, M, Fuchs, J, Demmer, U. | | Deposit date: | 2024-02-16 | | Release date: | 2025-03-05 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Enzymatic Birch reduction via hydrogen atom transfer at [4Fe-4S]-OH 2 and [8Fe-9S] clusters.
Nat Commun, 16, 2025
|
|
8ID7
 
 | |
7CCW
 
 | | Crystal structure of death-associated protein kinase 1 in complex with resveratrol and MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Death-associated protein kinase 1, RESVERATROL, ... | | Authors: | Yokoyama, T, Suzuki, R, Mizuguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of death-associated protein kinase 1 in complex with the dietary compound resveratrol.
Iucrj, 8, 2020
|
|
8B7H
 
 | |
6U4W
 
 | | 1.4 A structure of a pathogenic human Syt 1 C2B (D366E) | | Descriptor: | SULFATE ION, Synaptotagmin-1 | | Authors: | Dominguez, M.J, Bradberry, M.M, Chapman, E.R, Sutton, R.B. | | Deposit date: | 2019-08-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis for Synaptotagmin-1-Associated Neurodevelopmental Disorder.
Neuron, 107, 2020
|
|
8J7A
 
 | | Coordinates of Cryo-EM structure of the Arabidopsis thaliana PSI in state 1 (PSI-ST1) | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
5HYU
 
 | |
6IX6
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase with N-propanol at 1.43 A resolution | | Descriptor: | N-PROPANOL, Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of the complex of peptidyl-tRNA hydrolase with N-propanol at 1.43 A resolution
To Be Published
|
|
5OKT
 
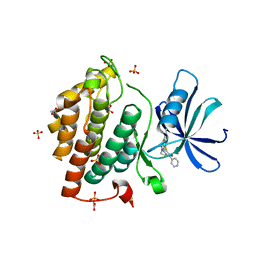 | | Crystal structure of human Casein Kinase I delta in complex with IWP-2 | | Descriptor: | ACETATE ION, Casein kinase I isoform delta, GLYCEROL, ... | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of Inhibitor of Wnt Production 2 (IWP-2) and Related Compounds As Selective ATP-Competitive Inhibitors of Casein Kinase 1 (CK1) delta / epsilon.
J. Med. Chem., 61, 2018
|
|
6JAW
 
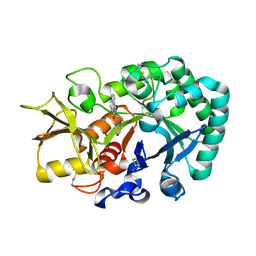 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a napthalimide derivative | | Descriptor: | 2-[3-(morpholin-4-yl)propyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAV
 
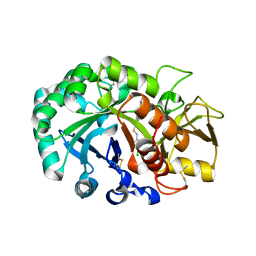 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a piperidine-thienopyridine derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-chlorophenyl)methyl]sulfanyl}-7-methyl-N-(prop-2-en-1-yl)-7,8-dihydropyrido[4',3':4,5]thieno[2,3-d]pyrimidin-4-amine, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.437 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6H5C
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 1 | | Descriptor: | (1~{S},3~{R},4~{S},5~{R})-3-methyl-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic Acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
7CJV
 
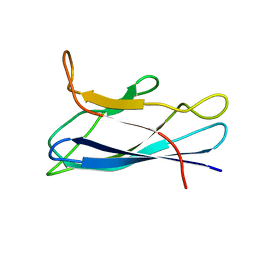 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a dilute environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
5VIS
 
 | | 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil Uncultured Bacterium. | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, Dihydropteroate Synthase, ... | | Authors: | Minasov, G, Wawrzak, Z, Di Leo, R, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-17 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil
Uncultured Bacterium.
To Be Published
|
|
5N51
 
 | |
5ZJP
 
 | |
6RT1
 
 | | Native tetragonal lysozyme - home source data | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Pereira, P.J.B. | | Deposit date: | 2019-05-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.336 Å) | | Cite: | Protein crystals as a key for deciphering macromolecular crowding effects on biological reactions.
Phys Chem Chem Phys, 22, 2020
|
|
6RT3
 
 | | Native tetragonal lysozyme - synchrotron data | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Pereira, P.J.B. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Protein crystals as a key for deciphering macromolecular crowding effects on biological reactions.
Phys Chem Chem Phys, 22, 2020
|
|
5K4W
 
 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NADH and L-threonine refined to 1.72 angstroms | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-threonine 3-dehydrogenase, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
9DL2
 
 | | Structure of proline utilization A complexed with 2,3-dihydro-1,4-benzodioxine-5-carboxylic acid | | Descriptor: | 2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2024-09-10 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystallographic Fragment Screening of a Bifunctional Proline Catabolic Enzyme Reveals New Inhibitor Templates for Proline Dehydrogenase and L-Glutamate-gamma-semialdehyde Dehydrogenase.
Molecules, 29, 2024
|
|
