4IBA
 
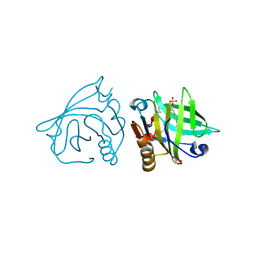 | | Bovine beta-lactoglobulin (isoform B) in complex with dodecyl sulphate (SDS) | | Descriptor: | DODECYL SULFATE, GLYCEROL, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
3R1L
 
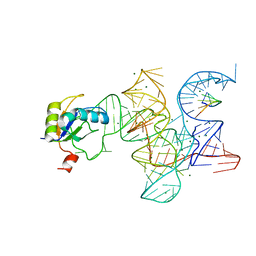 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Mg2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', Class I ligase ribozyme, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4D8D
 
 | |
4TZ4
 
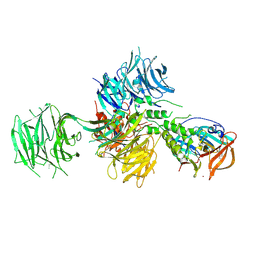 | | Crystal Structure of Human Cereblon in Complex with DDB1 and Lenalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Lenalidomide, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B, Riley, M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
To Be Published
|
|
2Z7K
 
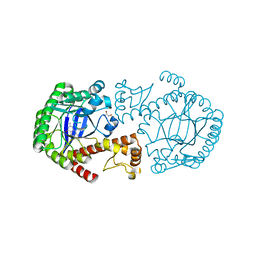 | | tRNA-Guanine transglycosylase (TGT) in complex with 2-Amino-lin-Benzoguanine | | Descriptor: | 2,6-diamino-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2007-08-24 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure analysis and in silico pKa calculations suggest strong pKa shifts of ligands as driving force for high-affinity binding to TGT
Chembiochem, 10, 2009
|
|
5MVN
 
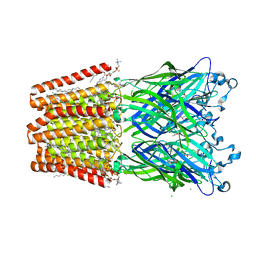 | |
7BR3
 
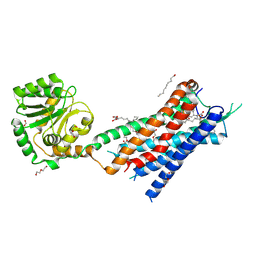 | | Crystal structure of the protein 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | Authors: | Cheng, L, Shao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
4X6C
 
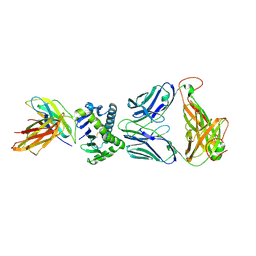 | | CD1a ternary complex with lysophosphatidylcholine and BK6 TCR | | Descriptor: | (4R,7R,18Z)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
5MUO
 
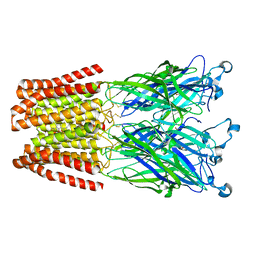 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Fourati, Z, Ruza, R.R, Delarue, M. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
4X6B
 
 | | BK6 TCR apo structure | | Descriptor: | TCR alpha, TCR beta | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-07 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
5MZQ
 
 | | X-ray structure of the M205W mutant of GLIC in complex with bromoform | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, ... | | Authors: | Fourati, Z, Delarue, M. | | Deposit date: | 2017-02-01 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
6T5W
 
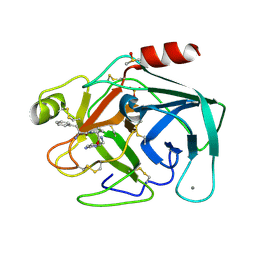 | | Cationic Trypsin in Complex with a D-Phe-Pro-p-aminopyridine Derivative (cocrystallizaton at 291 K) | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(6-azanylpyridin-3-yl)methyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic Trypsin, ... | | Authors: | Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-10-17 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Protein-Induced Change in Ligand Protonation during Trypsin and Thrombin Binding: Hint on Differences in Selectivity Determinants of Both Proteins?
J.Med.Chem., 63, 2020
|
|
5XF9
 
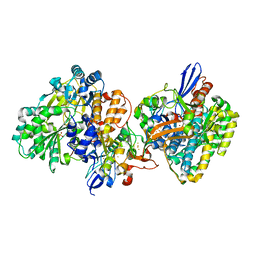 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
4X6D
 
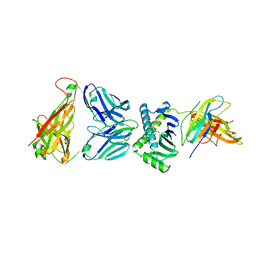 | | CD1a ternary complex with endogenous lipids and BK6 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, OLEIC ACID, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
3DKM
 
 | | Crystal structure of the HECTD1 CPH domain | | Descriptor: | E3 ubiquitin-protein ligase HECTD1 | | Authors: | Walker, J.R, Qiu, L, Li, Y, Bountra, C, Wolkstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-25 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the CPH domain of the E3 ubiquitin-protein ligase HECTD1.
To be Published
|
|
6F5R
 
 | | Crystal Structure of KDM4D with GF028 ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-oxidanylpropylamino)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.607 Å) | | Cite: | Crystal Structure of KDM4D with GF028 ligand
To be published
|
|
6T4A
 
 | | Thrombin in Complex with a D-Phe-Pro-p-aminopyridine derivative | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(6-azanylpyridin-3-yl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Ngo, K, Collins, C, Heine, A, Klebe, G. | | Deposit date: | 2019-10-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Protein-Induced Change in Ligand Protonation during Trypsin and Thrombin Binding: Hint on Differences in Selectivity Determinants of Both Proteins?
J.Med.Chem., 63, 2020
|
|
6F5S
 
 | | Crystal Structure of KDM4D with tetrazole ligand GF049 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of KDM4D with tetrazole ligand GF049
To be published
|
|
6TD8
 
 | | Human Aldose Reductase Mutant L301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid) | | Descriptor: | Aldo-keto reductase family 1 member B1, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Human Aldose Reductase Mutant L301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid)
To Be Published
|
|
7PQT
 
 | | Apo human Kv3.1 cryo-EM structure | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
6FAL
 
 | |
6TTK
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
9CB3
 
 | | E2F1-Cyclin F Interface | | Descriptor: | Cyclin-F, E2F1 peptide, S-phase kinase-associated protein 1 | | Authors: | Ngoi, P, Serrao, V.H, Rubin, S.M. | | Deposit date: | 2024-06-18 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural mechanism for the recognition of E2F1 by the ubiquitin ligase adaptor Cyclin F.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1QL9
 
 | |
6M94
 
 | | Monophosphorylated pSer33 b-Catenin peptide bound to b-TrCP/Skp1 Complex | | Descriptor: | Catenin beta-1, DIMETHYL SULFOXIDE, F-box/WD repeat-containing protein 1A, ... | | Authors: | Simonetta, K.R, Clifton, M.C, Walter, R.L, Ranieri, G.M, Carter, J.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction.
Nat Commun, 10, 2019
|
|
