7E75
 
 | |
7LVI
 
 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND (2R)-2-AMINO-N-[3-METHOXY-4- (1,3-OXAZOL-5-YL)PHENYL]-4-METHYLPENTANAMIDE | | Descriptor: | (2R,3R)-N-[3-methoxy-4-(1,3-oxazol-5-yl)phenyl]-3-(propan-2-yl)piperidine-2-carboxamide, 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery, Structure-Activity Relationships, and In Vivo Evaluation of Novel Aryl Amides as Brain Penetrant Adaptor Protein 2-Associated Kinase 1 (AAK1) Inhibitors for the Treatment of Neuropathic Pain.
J.Med.Chem., 64, 2021
|
|
7LVH
 
 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND N-[3-METHOXY-4-(1,3-OXAZOL-5-YL)PHENYL]-3-(PROPAN-2-YL)PIPERIDINE-2-CARBOXAMIDE | | Descriptor: | 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, N-[3-methoxy-4-(1,3-oxazol-5-yl)phenyl]-D-leucinamide, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery, Structure-Activity Relationships, and In Vivo Evaluation of Novel Aryl Amides as Brain Penetrant Adaptor Protein 2-Associated Kinase 1 (AAK1) Inhibitors for the Treatment of Neuropathic Pain.
J.Med.Chem., 64, 2021
|
|
7E6T
 
 | | Structural insights into the activation of human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYCLOMETHYLTRYPTOPHAN, ... | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
7E6U
 
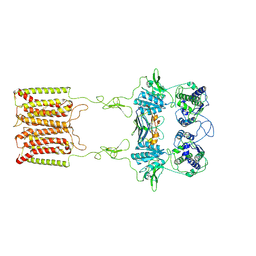 | | the complex of inactive CaSR and NB2D11 | | Descriptor: | Extracellular calcium-sensing receptor, NB-2D11 | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
7LV3
 
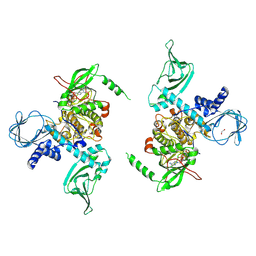 | | Crystal structure of human protein kinase G (PKG) R-C complex in inhibited state | | Descriptor: | 1,2-ETHANEDIOL, Isoform Beta of cGMP-dependent protein kinase 1, MANGANESE (II) ION, ... | | Authors: | Sharma, R, Lying, Q, Casteel, D, Kim, C. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | An auto-inhibited state of protein kinase G and implications for selective activation.
Elife, 11, 2022
|
|
7NMN
 
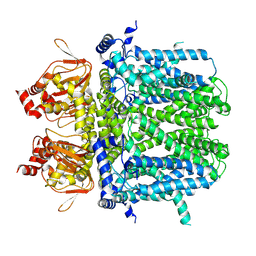 | | Rabbit HCN4 stabilised in amphipol A8-35 | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Rabbit HCN4 | | Authors: | Chaves-Sanjuan, A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7E6Q
 
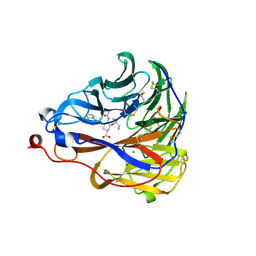 | | Crystal structure of influenza A virus neuraminidase N5 complexed with 4'-phenyl-1,2,3-triazolylated oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-acetamido-3-pentan-3-yloxy-5-(4-phenyl-1,2,3-triazol-1-yl)cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, P.F, Babayemi, O.O, Li, C.N, Fu, L.F, Zhang, S.S, Qi, J.X, Lv, X, Li, X.B. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of 5'-substituted 1,2,3-triazolylated oseltamivir derivatives as potent influenza neuraminidase inhibitors.
Rsc Adv, 11, 2021
|
|
7NL6
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | CALCIUM ION, DC-SIGN, CRD domain, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7NL7
 
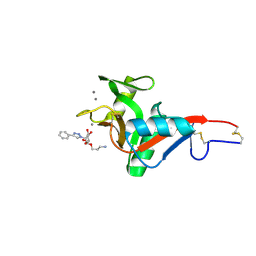 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | 3-Aminopropyl 2-deoxy-2-(4-phenyl-1,2,3-triazol-1-yl)-alpha-D-mannopyranoside, CALCIUM ION, DC-SIGN, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7NL9
 
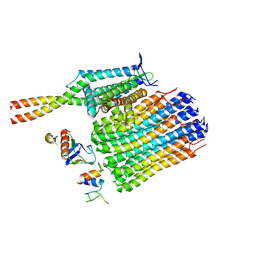 | | Mycobacterium smegmatis ATP synthase Fo state 3 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E6J
 
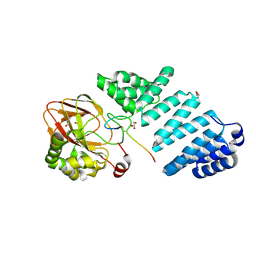 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) H725A in complex with Factor X peptide fragment (39mer-4Ser) | | Descriptor: | Aspartyl/asparaginyl beta-hydroxylase, GLYCEROL, PROPANOIC ACID, ... | | Authors: | Nakashima, Y, Brasnett, A, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Oxygenase Variants Employing a Single Protein Fe II Ligand Are Catalytically Active.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LTV
 
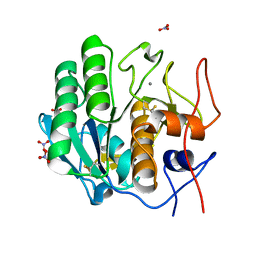 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 3 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU0
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 4 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU1
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 5 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU2
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 6 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU3
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 7 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LTD
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 1 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LTI
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 2 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7NKU
 
 | | diazaborine bound Drg1(AFG2) | | Descriptor: | 6-METHYL-2(PROPANE-1-SULFONYL)-2H-THIENO[3,2-D][1,2,3]DIAZABORININ-1-OL, ATPase family gene 2 protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Prattes, M, Bergler, H, Haselbach, D. | | Deposit date: | 2021-02-19 | | Release date: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inhibition of the AAA-ATPase Drg1 by diazaborine.
Nat Commun, 12, 2021
|
|
7NKW
 
 | | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-02-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
7E5N
 
 | | crystal structure of cis assembled TROP-2 | | Descriptor: | Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
7E5M
 
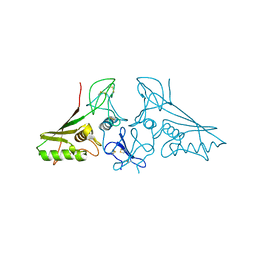 | | crystal structure of trans assembled human TROP-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
7LSQ
 
 | |
7LSK
 
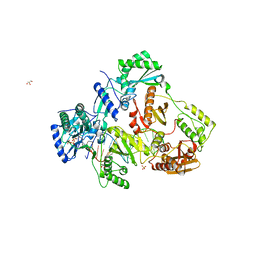 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, L-dTTP, and CA(2+) ion | | Descriptor: | 1-{2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-L-erythro-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-18 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
