6OLP
 
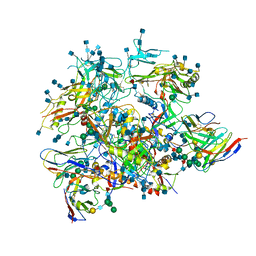 | | Full length HIV-1 Env AMC011 in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rantalainen, K, Cottrell, C.A. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics.
Plos Pathog., 15, 2019
|
|
5UM8
 
 | | Crystal structure of HIV-1 envelope trimer 16055 NFL TD CC (T569G) in complex with Fabs 35022 and PGT124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 35022 heavy chain, ... | | Authors: | Garces, F, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.935 Å) | | Cite: | Glycine Substitution at Helix-to-Coil Transitions Facilitates the Structural Determination of a Stabilized Subtype C HIV Envelope Glycoprotein.
Immunity, 46, 2017
|
|
3J5M
 
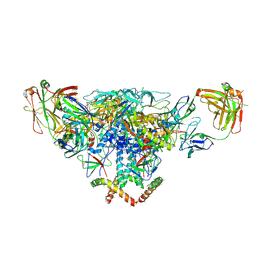 | | Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 3 PGV04 Fabs | | Descriptor: | BG505 SOSIP gp120, BG505 SOSIP gp41, PGV04 heavy chain, ... | | Authors: | Lyumkis, D, Julien, J.-P, Wilson, I.A, Ward, A.B. | | Deposit date: | 2013-10-26 | | Release date: | 2013-11-13 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
8G4P
 
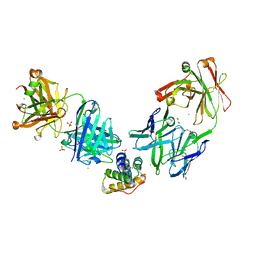 | | Crystal structure of the peanut allergen Ara h 2 bound by two neutralizing antibodies 13T1 and 13T5 | | Descriptor: | 1,2-ETHANEDIOL, 13T1 Fab light chain, 13T5 Fab heavy chain, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Min, J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design of an Ara h 2 hypoallergen from conformational epitopes.
Clin Exp Allergy, 54, 2024
|
|
5WK4
 
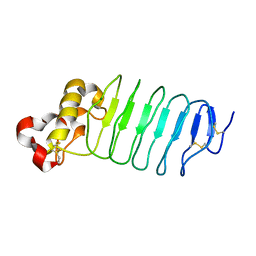 | | Crystal structure of an anti-idiotype VLR | | Descriptor: | MAGNESIUM ION, Variable lymphocyte receptor 39 | | Authors: | Collins, B.C, Nakahara, H, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-07-24 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystal structure of an anti-idiotype variable lymphocyte receptor.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5E0Q
 
 | |
4GKO
 
 | | Crystal structure of the calcium2+-bound human IgE-Fc(epsilon)3-4 bound to its B cell receptor derCD23 | | Descriptor: | CALCIUM ION, Ig epsilon chain C region, Low affinity immunoglobulin epsilon Fc receptor, ... | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
8TGO
 
 | | Crystal structure of the BG505 triple tandem trimer gp140 HIV-1 Env in complex with PGT124 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 scFv, ... | | Authors: | Xian, Y, Yuan, M, Wilson, I.A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines.
Npj Vaccines, 9, 2024
|
|
4GT7
 
 | | An engineered disulfide bond reversibly traps the IgE-Fc3-4 in a closed, non-receptor binding conformation | | Descriptor: | Ig epsilon chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wurzburg, B.A, Kim, B.K, Jardetzky, T.S. | | Deposit date: | 2012-08-28 | | Release date: | 2012-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An engineered disulfide bond reversibly traps the IgE-Fc3-4 in a closed, nonreceptor binding conformation.
J.Biol.Chem., 287, 2012
|
|
7SEG
 
 | | Crystal structure of the complex of CD16A bound by an anti-CD16A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Kiefer, J.R, Wallweber, H.A, Polson, A.G. | | Deposit date: | 2021-09-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | A BCMA/CD16A bispecific innate cell engager for the treatment of multiple myeloma.
Leukemia, 36, 2022
|
|
4J6K
 
 | |
4J6P
 
 | |
4J6L
 
 | |
4JPI
 
 | | Crystal structure of a putative VRC01 germline precursor Fab | | Descriptor: | GLYCEROL, Putative VRC01 germline Fab heavy chain, Putative VRC01 germline Fab light chain | | Authors: | Julien, J.-P, Diwanji, D.C, Jardine, J, Schief, W.R, Wilson, I.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational HIV immunogen design to target specific germline B cell receptors.
Science, 340, 2013
|
|
4J6Q
 
 | |
4J6N
 
 | |
4J6J
 
 | |
4J6M
 
 | |
6D2P
 
 | |
4NCO
 
 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP gp120, BG505 SOSIP gp41, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Lyumkis, D, Ward, A.B, Wilson, I.A. | | Deposit date: | 2013-10-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Crystal structure of a soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
7E7Y
 
 | |
7E7X
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E88
 
 | |
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
