6BD8
 
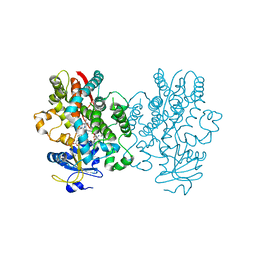 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, S-{(2S)-2-[(1-tert-butoxyethenyl)amino]-3-phenylpropyl}-N~2~-cyclopentyl-N-[(pyridin-3-yl)methyl]-L-cysteinamide | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-21 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Interaction of the rationally designed ritonavir-like inhibitors with human cytochrome P450 3A4: Impact of the side group interplay
Mol. Pharm., 2017
|
|
6BDK
 
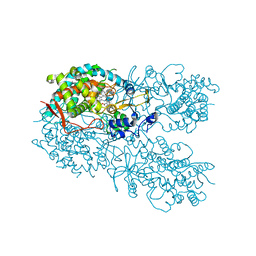 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2R)-2-(cyclopentylamino)-3-oxo-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}-3-(1H-indol-3-yl)propan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-23 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Inhibition of Human CYP3A4 by Rationally Designed Ritonavir-Like Compounds: Impact and Interplay of the Side Group Functionalities.
Mol Pharm., 15, 2018
|
|
6ZQD
 
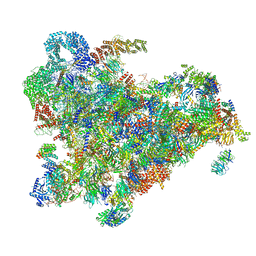 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Post-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQG
 
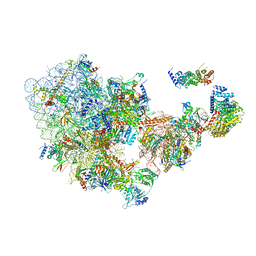 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-C | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6BD7
 
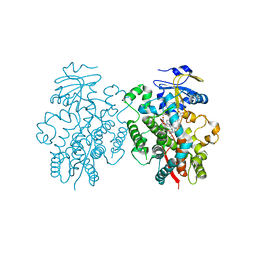 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2R)-3-oxo-2-[(propan-2-yl)amino]-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-21 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Interaction of the rationally designed ritonavir-like inhibitors with human cytochrome P450 3A4: Impact of the side group interplay
Mol. Pharm., 2017
|
|
1JWK
 
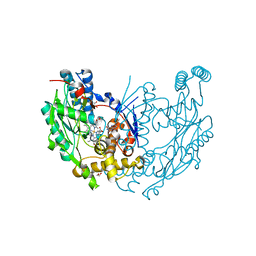 | | Murine Inducible Nitric Oxide Synthase Oxygenase Dimer (Delta 65) with W457A Mutation at Tetrahydrobiopterin Binding Site | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Aoyagi, M, Arvai, A.S, Ghosh, S, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of tetrahydrobiopterin binding-site mutants of inducible nitric oxide synthase oxygenase dimer and implicated roles of Trp457.
Biochemistry, 40, 2001
|
|
1JLI
 
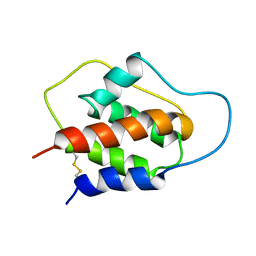 | |
2W13
 
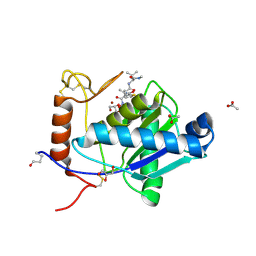 | | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | | Descriptor: | (2R,3R)-N^1^-[(1S)-2,2-DIMETHYL-1-(METHYLCARBAMOYL)PROPYL]-N^4^-HYDROXY-2-(2-METHYLPROPYL)-3-{[(1,3-THIAZOL-2-YLCARBONYL)AMINO]METHYL}BUTANEDIAMIDE, ACETATE ION, GLYCEROL, ... | | Authors: | Lingott, T.J, Schleberger, C, Gutierrez, J.M, Merfort, I. | | Deposit date: | 2008-10-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding.
Biochemistry, 48, 2009
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
2W12
 
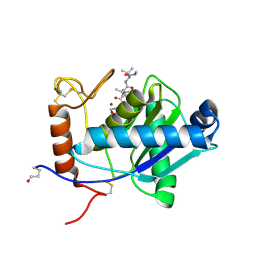 | | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | | Descriptor: | (2R,3R)-N^1^-[(1S)-2,2-DIMETHYL-1-(METHYLCARBAMOYL)PROPYL]-N^4^-HYDROXY-2-(2-METHYLPROPYL)-3-{[(1,3-THIAZOL-2-YLCARBONYL)AMINO]METHYL}BUTANEDIAMIDE, GLYCEROL, ZINC ION, ... | | Authors: | Lingott, T.J, Schleberger, C, Gutierrez, J.M, Merfort, I. | | Deposit date: | 2008-10-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding.
Biochemistry, 48, 2009
|
|
2W6Z
 
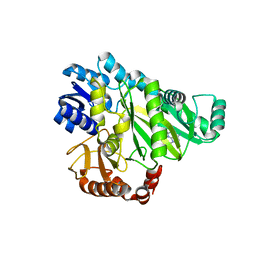 | |
2W14
 
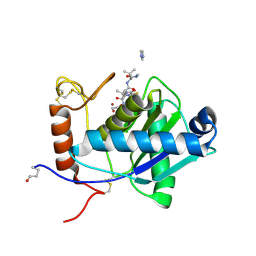 | | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | | Descriptor: | (2R,3R)-N^1^-[(1S)-2,2-DIMETHYL-1-(METHYLCARBAMOYL)PROPYL]-N^4^-HYDROXY-2-(2-METHYLPROPYL)-3-{[(1,3-THIAZOL-2-YLCARBONYL)AMINO]METHYL}BUTANEDIAMIDE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lingott, T.J, Schleberger, C, Gutierrez, J.M, Merfort, I. | | Deposit date: | 2008-10-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding.
Biochemistry, 48, 2009
|
|
2W6O
 
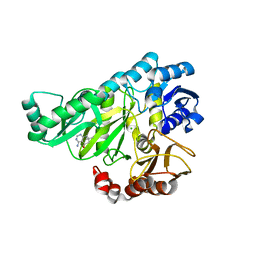 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | Descriptor: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
1K3B
 
 | | Crystal Structure of Human Dipeptidyl Peptidase I (Cathepsin C): Exclusion Domain Added to an Endopeptidase Framework Creates the Machine for Activation of Granular Serine Proteases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Turk, D, Janjic, V, Stern, I, Podobnik, M, Lamba, D, Dahl, S.W, Lauritzen, C, Pedersen, J, Turk, V, Turk, B. | | Deposit date: | 2001-10-02 | | Release date: | 2002-04-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases.
EMBO J., 20, 2001
|
|
2WEW
 
 | | Crystal structure of human apoM in complex with myristic acid | | Descriptor: | 1,2-ETHANEDIOL, APOLIPOPROTEIN M, MYRISTIC ACID | | Authors: | Sevvana, M, Ahnstrom, J, Egerer-Sieber, C, Dahlback, B, Muller, Y.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serendipitous Fatty Acid Binding Reveals the Structural Determinants for Ligand Recognition in Apolipoprotein M.
J.Mol.Biol., 393, 2009
|
|
2W9J
 
 | | The crystal structure of SRP14 from the Schizosaccharomyces pombe signal recognition particle | | Descriptor: | SIGNAL RECOGNITION PARTICLE SUBUNIT SRP14 | | Authors: | Brooks, M.A, Ravelli, R.B.G, McCarthy, A.A, Strub, K, Cusack, S. | | Deposit date: | 2009-01-25 | | Release date: | 2009-02-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Srp14 from the Schizosaccharomyces Pombe Signal Recognition Particle.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1KDO
 
 | | CYTIDINE MONOPHOSPHATE KINASE FROM E. COLI IN COMPLEX WITH CYTIDINE MONOPHOSPHATE | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, CYTIDYLATE KINASE, SULFATE ION | | Authors: | Bertrand, T, Briozzo, P, Assairi, L, Ofiteru, A, Bucurenci, N, Munier-Lehmann, H, Golinelli-Pimpaneau, B, Barzu, O, Gilles, A.M. | | Deposit date: | 2001-11-13 | | Release date: | 2002-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme.
J.Mol.Biol., 315, 2002
|
|
2W6P
 
 | |
2W6M
 
 | |
1KDR
 
 | | CYTIDINE MONOPHOSPHATE KINASE FROM E.COLI IN COMPLEX WITH ARA-CYTIDINE MONOPHOSPHATE | | Descriptor: | CYTIDYLATE KINASE, CYTOSINE ARABINOSE-5'-PHOSPHATE, SULFATE ION | | Authors: | Bertrand, T, Briozzo, P, Assairi, L, Ofiteru, A, Bucurenci, N, Munier-Lehmann, H, Golinelli-Pimpaneau, B, Barzu, O, Gilles, A.M. | | Deposit date: | 2001-11-13 | | Release date: | 2002-01-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme.
J.Mol.Biol., 315, 2002
|
|
1KV9
 
 | | Structure at 1.9 A Resolution of a Quinohemoprotein Alcohol Dehydrogenase from Pseudomonas putida HK5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETONE, CALCIUM ION, ... | | Authors: | Chen, Z.-W, Matsushita, K, Yamashita, T, Fujii, T, Toyama, H, Adachi, O, Bellamy, H.D, Mathews, F.S. | | Deposit date: | 2002-01-25 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Structure, 10, 2002
|
|
1KDT
 
 | | CYTIDINE MONOPHOSPHATE KINASE FROM E.COLI IN COMPLEX WITH 2',3'-DIDEOXY-CYTIDINE MONOPHOSPHATE | | Descriptor: | 2',3'-DIDEOXYCYTIDINE-5'-MONOPHOSPHATE, CYTIDYLATE KINASE, SULFATE ION | | Authors: | Bertrand, T, Briozzo, P, Assairi, L, Ofiteru, A, Bucurenci, N, Munier-Lehmann, H, Golinelli-Pimpaneau, B, Barzu, O, Gilles, A.M. | | Deposit date: | 2001-11-13 | | Release date: | 2002-01-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme.
J.Mol.Biol., 315, 2002
|
|
3CYU
 
 | | Human Carbonic Anhydrase II complexed with Cryptophane biosensor and xenon | | Descriptor: | Carbonic anhydrase 2, MoMo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-y l)propanoic acid]-cryptophane-A, PoPo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-yl)propanoic acid]-cryptophane-A, ... | | Authors: | Aaron, J.A, Jude, K.M, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2008-04-26 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a 129Xe-cryptophane biosensor complexed with human carbonic anhydrase II.
J.Am.Chem.Soc., 130, 2008
|
|
5VCC
 
 | | Crystal structure of human CYP3A4 bound to glycerol | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-03-31 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Level Production and Properties of the Cysteine-Depleted Cytochrome P450 3A4.
Biochemistry, 56, 2017
|
|
7NVR
 
 | | Human Mediator with RNA Polymerase II Pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Rengachari, S, Schilbach, S, Aibara, S, Cramer, P. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
