2LVK
 
 | | Solution structure of Ca-bound Phl p 7 | | Descriptor: | CALCIUM ION, Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Sirianni, A.G, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
7QZY
 
 | | ATAD2 in complex with FragLite29 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1-(2-methoxyethyl)pyridin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QYK
 
 | | ATAD2 in complex with FragLite18 | | Descriptor: | (4-bromanylpyridin-2-yl)methanol, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7CYB
 
 | | Saimiri boliviensis boliviensis galectin-13 with glycerol | | Descriptor: | GLYCEROL, Galectin | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
7CYA
 
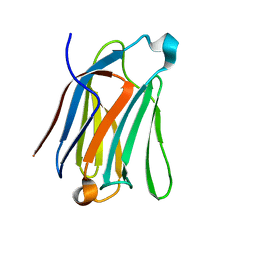 | | Saimiri boliviensis boliviensis galectin-13 with lactose | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
4JFT
 
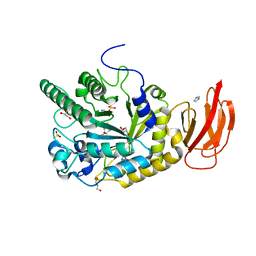 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor N-desmethyl-4-epi-(+)-Codonopsinine | | Descriptor: | (2S,3S,4R,5S)-2-(4-methoxyphenyl)-5-methylpyrrolidine-3,4-diol, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
5N5R
 
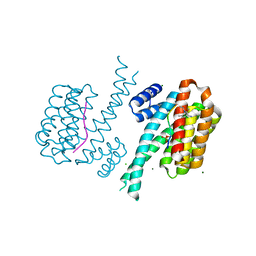 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV1 | | Descriptor: | 14-3-3 protein sigma, 2-azanyl-~{N}-(2,6-dimethylphenyl)-~{N}-propan-2-yl-ethanamide, CHLORIDE ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
5V89
 
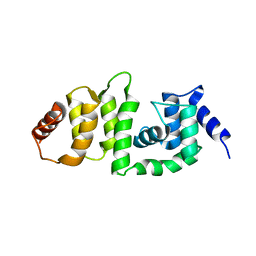 | | Structure of DCN4 PONY domain bound to CUL1 WHB | | Descriptor: | Cullin-1, DCN1-like protein 4 | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
2XXY
 
 | |
7VNH
 
 | |
3VB1
 
 | |
7VNA
 
 | | drosophlia AHR PAS-B domain | | Descriptor: | Ahr homolog spineless | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-10 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
5N75
 
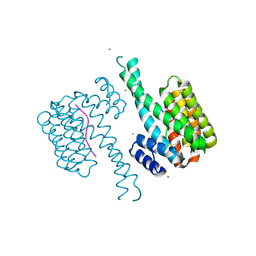 | | 14-3-3 sigma in complex with TAZ pS89 peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-19 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
4JFS
 
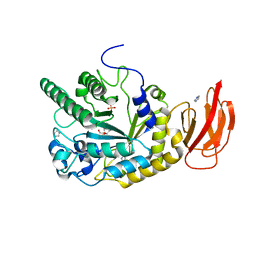 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor 4-epi-(+)-Codonopsinine | | Descriptor: | (2S,3S,4R,5S)-2-(4-methoxyphenyl)-1,5-dimethylpyrrolidine-3,4-diol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
6HM5
 
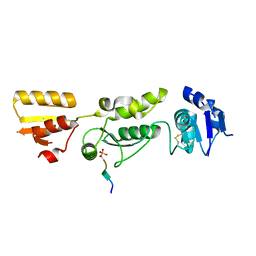 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a RAD9 phosphopeptide | | Descriptor: | Cell cycle checkpoint control protein RAD9A, DNA topoisomerase II binding protein 1 | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.330038 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
6GHV
 
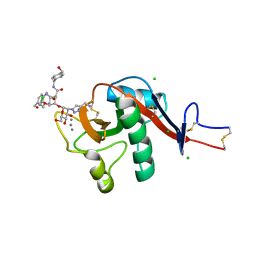 | | Structure of a DC-SIGN CRD in complex with high affinity glycomimetic. | | Descriptor: | CALCIUM ION, CD209 antigen, CHLORIDE ION, ... | | Authors: | Thepaut, M, Achilli, S, Medve, L, Bernardi, A, Fieschi, F. | | Deposit date: | 2018-05-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancing Potency and Selectivity of a DC-SIGN Glycomimetic Ligand by Fragment-Based Design: Structural Basis.
Chemistry, 25, 2019
|
|
3DCG
 
 | | Crystal Structure of the HIV Vif BC-box in Complex with Human ElonginB and ElonginC | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Stanley, B.J, Ehrlich, E.S, Short, L, Yu, Y, Xiao, Z, Yu, X.-F, Xiong, Y. | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly
J.Virol., 82, 2008
|
|
3V2L
 
 | |
6XVC
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 1 | | Descriptor: | (4~{R})-4-[(1~{R})-1-[7-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)quinolin-5-yl]oxyethyl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.098 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6HM3
 
 | | Crystal structure of Rad4 BRCT1,2 in complex with a Sld3 phosphopeptide | | Descriptor: | CALCIUM ION, DNA replication regulator sld3, GLYCEROL, ... | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77263618 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
4TZ4
 
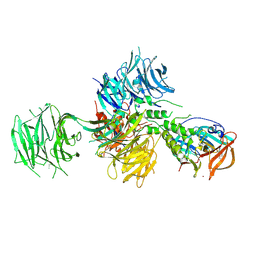 | | Crystal Structure of Human Cereblon in Complex with DDB1 and Lenalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Lenalidomide, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B, Riley, M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
To Be Published
|
|
8DLD
 
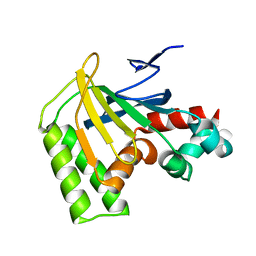 | | Crystal structure of chalcone-isomerase like protein from Physcomitrella patens (PpCHIL-A) | | Descriptor: | Chalcone-flavonone isomerase family protein | | Authors: | Wolf Saxon, E, Moorman, C, Castro, A, Ruiz, A, Mallari, J.P, Burke, J.R. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Regulatory ligand binding in plant chalcone isomerase-like (CHIL) proteins.
J.Biol.Chem., 299, 2023
|
|
8DLC
 
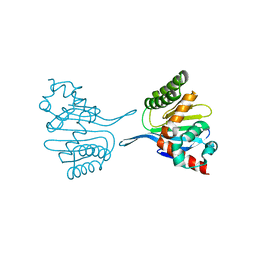 | | Crystal structure of chalcone-isomerase like protein from Vitis vinifera (VvCHIL) | | Descriptor: | Chalcone-flavonone isomerase family protein | | Authors: | Wolf Saxon, E, Moorman, C, Castro, A, Ruiz, A, Mallari, J.P, Burke, J.R. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulatory ligand binding in plant chalcone isomerase-like (CHIL) proteins.
J.Biol.Chem., 299, 2023
|
|
1F28
 
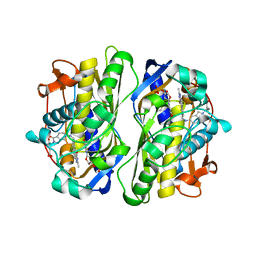 | | CRYSTAL STRUCTURE OF THYMIDYLATE SYNTHASE FROM PNEUMOCYSTIS CARINII BOUND TO DUMP AND BW1843U89 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Anderson, A.C, O'Neil, R.H, Surti, T.S, Stroud, R.M. | | Deposit date: | 2000-05-23 | | Release date: | 2001-06-06 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Approaches to solving the rigid receptor problem by identifying a minimal set of flexible residues during ligand docking.
Chem.Biol., 8, 2001
|
|
3R1H
 
 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Ca2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', CALCIUM ION, Class I ligase ribozyme, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
