3OBS
 
 | | Crystal structure of Tsg101 UEV domain | | Descriptor: | Tumor susceptibility gene 101 protein | | Authors: | Im, Y.J, Hurley, J.H. | | Deposit date: | 2010-08-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic and Functional Analysis of the ESCRT-I /HIV-1 Gag PTAP Interaction.
Structure, 18, 2010
|
|
4FQI
 
 | |
7E0J
 
 | | LHCII-1 in the state transition supercomplex PSI-LHCI-LHCII from the double phosphatase mutant pph1;pbcp of Chlamydomonas reinhardti. | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-06-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
3P7B
 
 | | p38 inhibitor-bound | | Descriptor: | 1-{5-tert-butyl-3-[(5-oxo-1,4-diazepan-1-yl)carbonyl]thiophen-2-yl}-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ONE
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6B3P
 
 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K in Complex with Maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, Amy13K, FORMIC ACID, ... | | Authors: | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
7E7Q
 
 | | Cryo-EM structure of human ABCA4 in ATP-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
4GK3
 
 | | Human EphA3 Kinase domain in complex with ligand 87 | | Descriptor: | 8-butyl-1-methyl-7-(2-methylphenyl)-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
6KYI
 
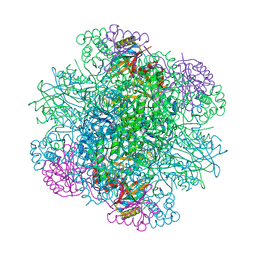 | | Rice Rubisco in complex with sulfate ions | | Descriptor: | GLYCEROL, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain, ... | | Authors: | Matsumura, H, Yoshizawa, T, Tanaka, S, Yoshikawa, H. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hybrid Rubisco with Complete Replacement of Rice Rubisco Small Subunits by Sorghum Counterparts Confers C 4 Plant-like High Catalytic Activity.
Mol Plant, 13, 2020
|
|
3OY0
 
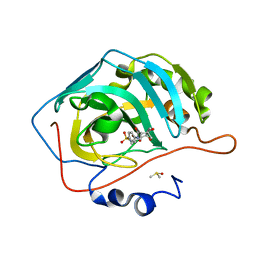 | | Human Carbonic Anhydrase II complexed with 1-(4-(4-(2-(ISOPROPYLSULFONYL)PHENYLAMINO)-1H-PYRROLO[2,3-B]PYRIDIN-6-YLAMINO)-3-METHOXYPHENYL)PIPERIDIN-4-OL | | Descriptor: | (2S)-2-tert-butyl-N-(4-sulfamoylphenyl)pentanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Aggarwal, M, McKenna, R. | | Deposit date: | 2010-09-22 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Anticonvulsant 4-aminobenzenesulfonamide derivatives with branched-alkylamide moieties: X-ray crystallography and inhibition studies of human carbonic anhydrase isoforms I, II, VII, and XIV.
J.Med.Chem., 54, 2011
|
|
2HXP
 
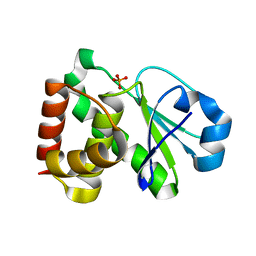 | | Crystal Structure of the human phosphatase (DUSP9) | | Descriptor: | Dual specificity protein phosphatase 9, PHOSPHATE ION | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
1WM2
 
 | |
2HY6
 
 | | A seven-helix coiled coil | | Descriptor: | General control protein GCN4, HEXANE-1,6-DIOL | | Authors: | Liu, J, Zheng, Q, Deng, Y, Cheng, C.S, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A seven-helix coiled coil.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4GK4
 
 | | Human EphA3 Kinase domain in complex with ligand 90 | | Descriptor: | 8-butyl-1-methyl-7-(5-methyl-1H-indazol-4-yl)-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
5QXI
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with PC587 | | Descriptor: | (4R,4aR,7aS,9R)-3,10-dimethyl-5,6,7,7a,8,9-hexahydro-4H-4a,9-epiminopyrrolo[3',4':5,6]cyclohepta[1,2-d][1,2]oxazol-4-ol, (4R,4aS,7aS,9S)-3,10-dimethyl-5,6,7,7a,8,9-hexahydro-4H-4a,9-epiminopyrrolo[3',4':5,6]cyclohepta[1,2-d][1,2]oxazol-4-ol, 1,2-ETHANEDIOL, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
2C5N
 
 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, N-[4-(2,4-DIMETHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YL]-N',N'-DIMETHYL-BENZENE-1,4-DIAMINE | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-10-30 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
2BIL
 
 | | The human protein kinase Pim1 in complex with its consensus peptide Pimtide | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, CONSENSUS PIM1 PEPTIDE PIMTIDE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Knapp, S, Debreczeni, J, Bullock, A, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Guo, K. | | Deposit date: | 2005-01-22 | | Release date: | 2005-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Human Protein Kinase Pim1 in Complex with its Consensus Peptide Pimtide
To be Published
|
|
4RFM
 
 | | ITK kinase domain in complex with compound 1 N-{1-[(1,1-dioxo-1-thian-2-yl)(phenyl)methyl]-1H- pyrazol-4-yl}-5,5-difluoro-5a-methyl-1H,4H,4aH,5H,5aH,6H-cyclopropa[f]indazole-3-carboxamide | | Descriptor: | (4aS,5aR)-N-{1-[(R)-[(2R)-1,1-dioxidotetrahydro-2H-thiopyran-2-yl](phenyl)methyl]-1H-pyrazol-4-yl}-5,5-difluoro-5a-methyl-1,4,4a,5,5a,6-hexahydrocyclopropa[f]indazole-3-carboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | McEwan, P.A, Barker, J.J, Eigenbrot, C. | | Deposit date: | 2014-09-26 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetrahydroindazoles as Interleukin-2 Inducible T-Cell Kinase Inhibitors. Part II. Second-Generation Analogues with Enhanced Potency, Selectivity, and Pharmacodynamic Modulation in Vivo.
J.Med.Chem., 58, 2015
|
|
2G00
 
 | | Factor Xa in complex with the inhibitor 3-(6-(2'-((dimethylamino)methyl)-4-biphenylyl)-7-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-c]pyridin-1-yl)benzamide | | Descriptor: | 3-[6-{2'-[(DIMETHYLAMINO)METHYL]BIPHENYL-4-YL}-7-OXO-3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN-1-YL]BENZAMIDE, Coagulation factor X | | Authors: | Alexander, R.S. | | Deposit date: | 2006-02-10 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of potent, efficacious, and orally bioavailable inhibitors of blood coagulation factor Xa with neutral P1 moieties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5VBI
 
 | |
2G0W
 
 | |
1J0H
 
 | |
2OOW
 
 | | MIF Bound to a Fluorinated OXIM Derivative | | Descriptor: | 3-FLUORO-4-HYDROXYBENZALDEHYDE O-(CYCLOHEXYLCARBONYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
1ARM
 
 | | CARBOXYPEPTIDASE A WITH ZN REPLACED BY HG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, HG-CARBOXYPEPTIDASE A=ALPHA= (COX), ... | | Authors: | Greenblatt, H.M, Feinberg, H, Tucker, P.A, Shoham, G. | | Deposit date: | 1994-11-22 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Carboxypeptidase A: native, zinc-removed and mercury-replaced forms.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1X1J
 
 | | Crystal Structure of Xanthan Lyase (N194A) with a Substrate. | | Descriptor: | (4AR,6R,7S,8R,8AR)-8-((5R,6R)-3-CARBOXY-TETRAHYDRO-4,5,6-TRIHYDROXY-2H-PYRAN-2-YLOXY)-HEXAHYDRO-6,7-DIHYDROXY-2-METHYLPYRANO[3,2-D][1,3]DIOXINE-2-CARBOXYLIC ACID), CALCIUM ION, xanthan lyase | | Authors: | Maruyama, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-04-04 | | Release date: | 2005-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase Complexed with a Substrate: Insights into the Enzyme Reaction Mechanism
J.Mol.Biol., 350, 2005
|
|
