4K8K
 
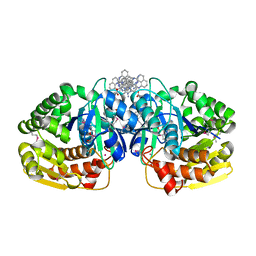 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 1-(4-methoxyphenyl)-1-cyclopropane and 2-aminoperimidine | | Descriptor: | 1-(4-methoxyphenyl)cyclopropanecarboxylic acid, 1H-perimidin-2-amine, ADENOSINE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 1-(4-methoxyphenyl)-1-cyclopropane and 2-aminoperimidine
To be Published
|
|
6WG7
 
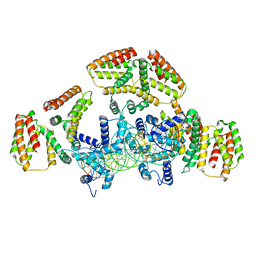 | | Coordinates of NanR dimer fitted in Hexameric NanR-DNA hetero-complex cryo-EM map | | Descriptor: | DNA (35-MER), HTH-type transcriptional repressor NanR | | Authors: | Hariprasad, V, Horne, C, Santosh, P, Amy, H, Emre, B, Rachel, N, Michael, G, Georg, R, Borries, D, Renwick, D. | | Deposit date: | 2020-04-05 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism.
Nat Commun, 12, 2021
|
|
7MNK
 
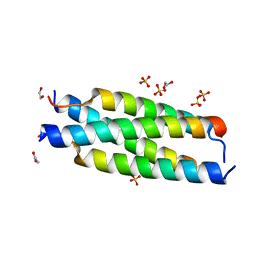 | | Crystal structure of the tetramerization element of NUP358/RanBP2 (residues 805-832) | | Descriptor: | 1,2-ETHANEDIOL, E3 SUMO-protein ligase RanBP2, SULFATE ION | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
4K93
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, N-(hydroxymethyl)benzamide, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE
To be Published
|
|
7PS0
 
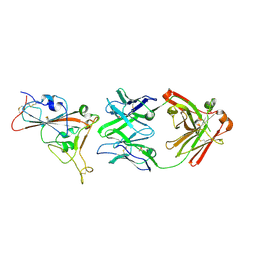 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with beta-24 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-24 heavy chain, Beta-24 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS4
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-38 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-38 Fab heavy chain, Beta-38 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
6QIL
 
 | | The complex structure of hsRosR-S1 (VNG0258H/RosR-S1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (28-MER), DNA binding protein, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-21 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
5W6J
 
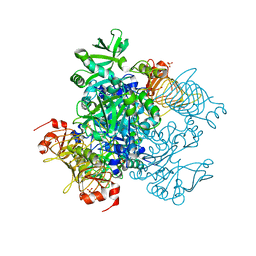 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Wu, R, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
1EET
 
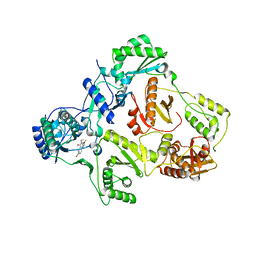 | | HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH THE INHIBITOR MSC204 | | Descriptor: | 1-(5-BROMO-PYRIDIN-2-YL)-3-[2-(6-FLUORO-2-HYDROXY-3-PROPIONYL-PHENYL)-CYCLOPROPYL]-UREA, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Hogberg, M, Sahlberg, C, Engelhardt, P, Noreen, R, Kangasmetsa, J, Johansson, N.G, Oberg, B, Vrang, L, Zhang, H, Sahlberg, B.L, Unge, T, Lovgren, S, Fridborg, K, Backbro, K. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues.
J.Med.Chem., 42, 1999
|
|
4UBC
 
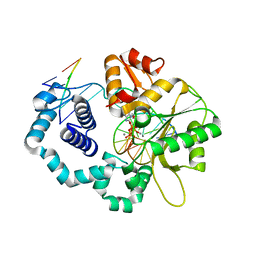 | | DNA polymerase beta substrate complex with a templating cytosine and incoming 8-oxodGTP, 0 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
1EON
 
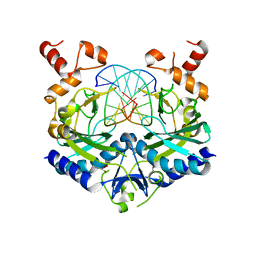 | | ECORV BOUND TO 3'-S-PHOSPHOROTHIOLATE DNA AND CA2+ | | Descriptor: | ACETIC ACID, CHLORIDE ION, DNA (5'-D(*AP*AP*AP*GP*AP*(TSP)P*AP*TP*CP*TP*T)-3'), ... | | Authors: | Horton, N.C, Connolly, B.A, Perona, J.J. | | Deposit date: | 2000-03-23 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of EcoRV Endonuclease by Deoxyribo-3'-S-phosphorothiolates: A High-Resolution X-ray Crystallographic Study
J.Am.Chem.Soc., 122, 2000
|
|
5WAE
 
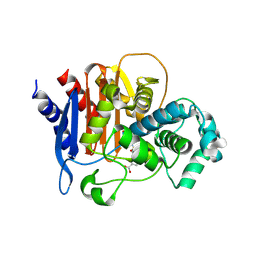 | | ADC-7 in complex with boronic acid transition state inhibitor CR167 | | Descriptor: | 3-(5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl)benzoic acid, Beta-lactamase | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
4K9C
 
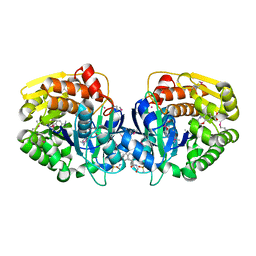 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE and 4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-CARBOXYLIC ACID | | Descriptor: | 4-methyl-3,4-dihydro-2H-1,4-benzoxazine-7-carboxylic acid, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE and 4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-CARBOXYLIC ACID
To be Published
|
|
1E56
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the natural substrate DIMBOA-beta-D-glucoside | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase- Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4TZ8
 
 | | Structure of human ATAD2 bromodomain bound to fragment inhibitor | | Descriptor: | 2-amino-7,7-dimethyl-5,6,7,8-tetrahydro-4H-[1,3]thiazolo[5,4-c]azepin-4-one, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-09 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
7MVS
 
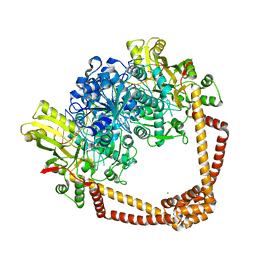 | | DNA gyrase complexed with uncleaved DNA and Compound 7 to 2.6A resolution | | Descriptor: | (1S)-2-[(2r,5S)-5-{[(2,3-dihydro-1,4-benzodioxin-6-yl)methyl]amino}-1,3-dioxan-2-yl]-1-(3-fluoro-6-methoxyquinolin-4-yl)ethan-1-ol, CHLORIDE ION, DNA (5'-D(P*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Ratigan, S.C, McElroy, C.A. | | Deposit date: | 2021-05-15 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60137153 Å) | | Cite: | Optimization of TopoIV Potency, ADMET Properties, and hERG Inhibition of 5-Amino-1,3-dioxane-Linked Novel Bacterial Topoisomerase Inhibitors: Identification of a Lead with In Vivo Efficacy against MRSA.
J.Med.Chem., 64, 2021
|
|
4U81
 
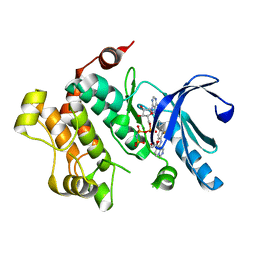 | | MEK1 Kinase bound to small molecule inhibitor G659 | | Descriptor: | 8-[(4-cyclopropyl-2-fluorophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-7-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Robarge, K.D, Ultsch, M.H, Weismann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1EPL
 
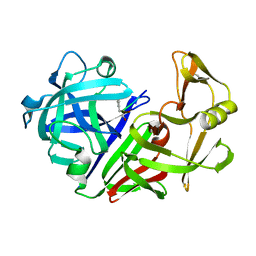 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS1, PRO-LEU-GLU-PSA-ARG-LEU | | Authors: | Al-Karadaghi, S, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EQH
 
 | | THE 2.7 ANGSTROM MODEL OF OVINE COX-1 COMPLEXED WITH FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, PROSTAGLANDIN H2 SYNTHASE-1, ... | | Authors: | Loll, P.J, Selinsky, B.S, Gupta, K, Sharkey, C.T. | | Deposit date: | 2000-04-04 | | Release date: | 2001-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations.
Biochemistry, 40, 2001
|
|
4AWY
 
 | | Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, METALLO-BETA-LACTAMASE AIM-1, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157
Antimicrob.Agents Chemother., 56, 2012
|
|
4UC0
 
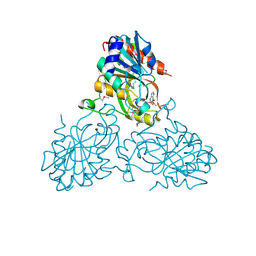 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
4UE3
 
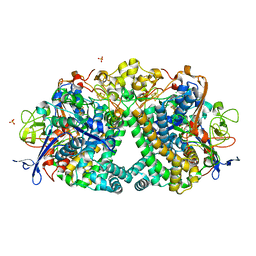 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases and the Importance of the active site Arginine | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
4U3H
 
 | | Crystal structure of FN3con | | Descriptor: | FN3con | | Authors: | Porebski, B.T, McGowan, S, Buckle, A.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and dynamic properties that govern the stability of an engineered fibronectin type III domain.
Protein Eng.Des.Sel., 28, 2015
|
|
5WBG
 
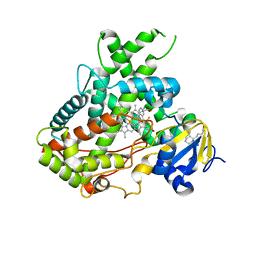 | | Crystal Structure of human Cytochrome P450 2B6 (Y226H/K262R) in complex with an analog of a drug Efavirenz | | Descriptor: | (2R,4S)-6-chloro-4-(cyclopropylethynyl)-2-methyl-4-(trifluoromethyl)-1,4-dihydro-2H-3,1-benzoxazine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, 5-cyclohexylpentan-1-ol, ... | | Authors: | Shah, M.B, Halpert, J.R. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog.
Int J Mol Sci, 19, 2018
|
|
4KAH
 
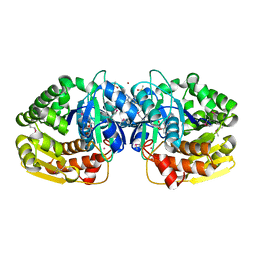 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 4-bromo-1H-pyrazole | | Descriptor: | 4-bromo-1H-pyrazole, ADENOSINE, BROMIDE ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 4-bromo-1H-pyrazole
To be Published
|
|
