4F6D
 
 | |
4O5O
 
 | |
4O5Y
 
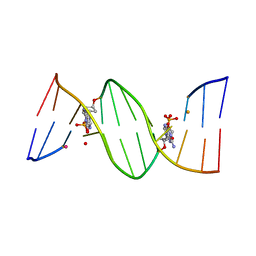 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), POTASSIUM ION | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4F8M
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23I/I92V at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Nam, S.P, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-05-17 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4NNK
 
 | |
4EZK
 
 | |
4NOZ
 
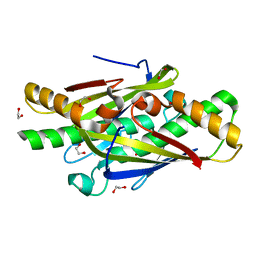 | | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Organic hydroperoxide resistance protein | | Authors: | Dranow, D.M, Lukacs, C.M, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia
TO BE PUBLISHED
|
|
4F1E
 
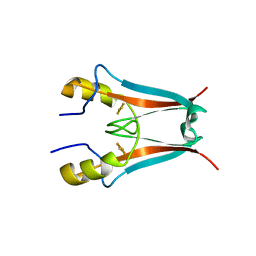 | | The Crystal Structure of a Human MitoNEET mutant with Asp 67 replaced by a Gly | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Baxter, E.I, Zuris, J.A, Wang, C, Axelrod, H.L, Cohen, A.E, Paddock, M.L, Nechushtai, R, Onuchic, J.N, Jennings, P.A. | | Deposit date: | 2012-05-06 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric control in a metalloprotein dramatically alters function.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4NVC
 
 | |
4NQ6
 
 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 complexed with compound L-CS319 | | Descriptor: | (3S,5S,7aR)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase 2, POTASSIUM ION, ... | | Authors: | Gonzalez, J.M, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4F45
 
 | | Crystal structure of human CD38 E226Q mutant in complex with NAADP | | Descriptor: | ADP-ribosyl cyclase 1, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2012-05-10 | | Release date: | 2013-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human CD38 in Complex with NAADP and ADPRP
MESSENGER, 2, 2013
|
|
4FK9
 
 | |
4NR7
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Krojer, T, Nowak, R, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
To be Published
|
|
4F6B
 
 | |
4NS1
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972.
To be Published
|
|
4EZL
 
 | |
4F09
 
 | |
4NTA
 
 | |
4F73
 
 | |
4O1N
 
 | |
4F7X
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4NVD
 
 | |
4NW3
 
 | | Crystal structure of MLL CXXC domain in complex with a CpG DNA | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*CP*GP*AP*TP*GP*GP*C)-3', Histone-lysine N-methyltransferase 2A, ZINC ION | | Authors: | Bian, C, Tempel, W, Chao, X, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
4NTB
 
 | |
4NWD
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | Descriptor: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
