1YNQ
 
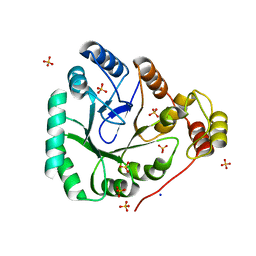 | | aldo-keto reductase AKR11C1 from Bacillus halodurans (holo form) | | Descriptor: | GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Marquardt, T, Kostrewa, D, Winkler, F.K, Li, X.D. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution Crystal Structure of AKR11C1 from Bacillus halodurans: An NADPH-dependent 4-Hydroxy-2,3-trans-nonenal Reductase
J.Mol.Biol., 354, 2005
|
|
2QBP
 
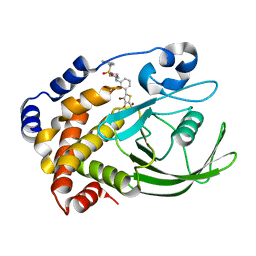 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
3IWO
 
 | |
3PVL
 
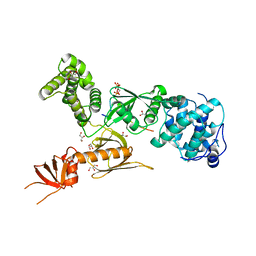 | | Structure of myosin VIIa MyTH4-FERM-SH3 in complex with the CEN1 of Sans | | Descriptor: | GLYCEROL, Myosin VIIa isoform 1, PHOSPHATE ION, ... | | Authors: | Wu, L, Pan, L.F, Wei, Z.Y, Zhang, M.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo.
Science, 331, 2011
|
|
3REO
 
 | | Monolignol O-methyltransferase (MOMT) | | Descriptor: | (Iso)eugenol O-methyltransferase, 2-methoxy-4-[(1E)-prop-1-en-1-yl]phenol, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bhuiya, M.W, Liu, C.J. | | Deposit date: | 2011-04-04 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monolignol O-methyltransferase (MOMT)
To be Published
|
|
4JUD
 
 | |
1UVC
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, STEARIC ACID | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
1JIJ
 
 | | Crystal structure of S. aureus TyrRS in complex with SB-239629 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]-(1,3,4,5-TETRAHYDROXY-4-HYDROXYMETHYL-PIPERIDIN-2-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
1JQ8
 
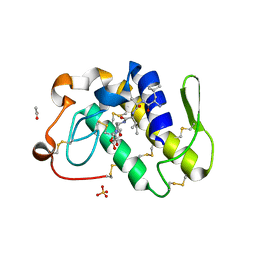 | | Design of specific inhibitors of phospholipase A2: Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Leu-Ala-Ile-Tyr-Ser at 2.0 resolution | | Descriptor: | ACETIC ACID, Peptide inhibitor, Phospholipase A2, ... | | Authors: | Chandra, V, Jasti, J, Kaur, P, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2001-08-04 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of specific peptide inhibitors of phospholipase A2: structure of a complex formed between Russell's viper phospholipase A2 and a designed peptide Leu-Ala-Ile-Tyr-Ser (LAIYS).
ACTA CRYSTALLOGR.,SECT.D, 58, 2002
|
|
2OHR
 
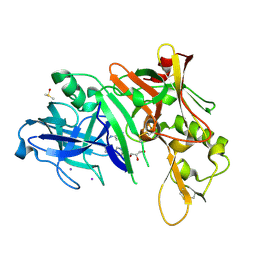 | | X-ray crystal structure of beta secretase complexed with compound 6a | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2UWO
 
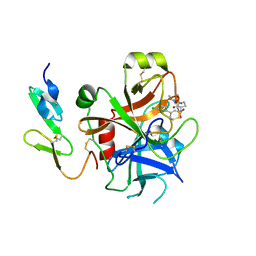 | | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa | | Descriptor: | (2R)-2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}PROPENE-1-SULFONAMIDE, COAGULATION FACTOR X | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5OHJ
 
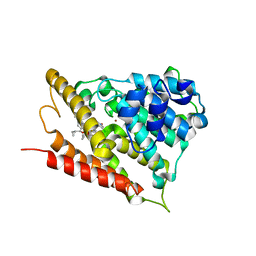 | | Human phosphodiesterase 4B catalytic domain in complex with a pyrrolidinyl inhibitor. | | Descriptor: | MAGNESIUM ION, ZINC ION, [(1~{S})-2-[3,5-bis(chloranyl)-1-oxidanyl-pyridin-1-ium-4-yl]-1-[4-[bis(fluoranyl)methoxy]-3-(cyclopropylmethoxy)phenyl]ethyl] (2~{S})-1-[3-(dimethylcarbamoyl)phenyl]sulfonylpyrrolidine-2-carboxylate, ... | | Authors: | Rizzi, A, Carzaniga, L, Armani, E. | | Deposit date: | 2017-07-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Thiazolidinyl and Pyrrolidinyl Derivatives as Inhaled PDE4 Inhibitors for Respiratory Diseases.
J. Med. Chem., 60, 2017
|
|
3SQQ
 
 | | CDK2 in complex with inhibitor RC-3-96 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-methylbenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-07-06 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
7XJR
 
 | | MLXase AlXyn26A | | Descriptor: | AlXyn26A | | Authors: | Zhang, Y.Z, Chen, X.L, Zhao, F, Yu, C.M. | | Deposit date: | 2022-04-18 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel class of xylanases specifically degrade marine red algal beta 1,3/1,4-mixed-linkage xylan.
J.Biol.Chem., 299, 2023
|
|
6K04
 
 | | Crystal structure of BRD2(BD2)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
1GKA
 
 | | The molecular basis of the coloration mechanism in lobster shell. beta-crustacyanin at 3.2 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASTAXANTHIN, ... | | Authors: | Cianci, M, Rizkallah, P.J, Olczak, A, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2001-08-10 | | Release date: | 2002-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | The Molecular Basis of the Coloration Mechanism in Lobster Shell: Beta -Crustacyanin at 3.2-A Resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2ZEB
 
 | | Potent, Nonpeptide Inhibitors of Human Mast Cell Tryptase | | Descriptor: | 1-(1'-{[3-(methylsulfanyl)-2-benzothiophen-1-yl]carbonyl}spiro[1-benzofuran-3,4'-piperidin]-5-yl)methanamine, Tryptase beta 2 | | Authors: | Spurlino, J.C, Lewandowski, F, Milligan, C. | | Deposit date: | 2007-12-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3R4N
 
 | | Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 4-[2-chloro-6-(4,4,4-trifluorobutoxy)phenyl]-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Gajiwala, K.S. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
4JHG
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with trans-zeatin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, MALONATE ION, MtN13 protein, ... | | Authors: | Ruszkowski, M, Tusnio, K, Ciesielska, A, Brzezinski, K, Dauter, M, Dauter, Z, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1BTN
 
 | | STRUCTURE OF THE BINDING SITE FOR INOSITOL PHOSPHATES IN A PH DOMAIN | | Descriptor: | BETA-SPECTRIN, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE | | Authors: | Wilmanns, M, Hyvoenen, M, Saraste, M. | | Deposit date: | 1995-08-23 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the binding site for inositol phosphates in a PH domain.
EMBO J., 14, 1995
|
|
4DAB
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with hypoxanthine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
2OGA
 
 | |
2ZLA
 
 | | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation and crystal structure | | Descriptor: | (1R,2S,3R,5Z,7E)-17-{(1R)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethoxy)-9,10-secoestra-5,7,16-triene-1,3-diol, Coactivator peptide DRIP, Vitamin D3 receptor | | Authors: | Shimizu, M, Miyamoto, Y, Nakabayashi, M, Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure
Bioorg.Med.Chem., 16, 2008
|
|
1OB0
 
 | | Kinetic stabilization of Bacillus licheniformis alpha-amylase through introduction of hydrophobic residues at the surface | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 2003-01-21 | | Release date: | 2003-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Kinetic Stabilization of Bacillus Licheniformis Alpha-Amylase Through Introduction of Hydrophobic Residues at the Surface
J.Biol.Chem., 278, 2003
|
|
4LDE
 
 | | Structure of beta2 adrenoceptor bound to BI167107 and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
