2QRC
 
 | | Crystal structure of the adenylate sensor from AMP-activated protein kinase in complex with ADP and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Protein C1556.08c, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
2OOX
 
 | |
4CYD
 
 | | GlxR bound to cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, PROBABLE EXPRESSION TAG, ... | | Authors: | Townsend, P.D, Bott, M, Cann, M.J, Pohl, E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structures of Apo and Camp-Bound Glxr from Corynebacterium Glutamicum Reveal Structural and Dynamic Changes Upon Camp Binding in Crp/Fnr Family Transcription Factors.
Plos One, 9, 2014
|
|
1ZPS
 
 | | Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI | | Descriptor: | ACETIC ACID, CADMIUM ION, Phosphoribosyl-AMP cyclohydrolase | | Authors: | Sivaraman, J, Myers, R.S, Boju, L, Sulea, T, Cygler, M, Davisson, V.J, Schrag, J.D, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Methanobacterium thermoautotrophicum Phosphoribosyl-AMP Cyclohydrolase HisI.
Biochemistry, 44, 2005
|
|
2E5A
 
 | | Crystal Structure of Bovine Lipoyltransferase in Complex with Lipoyl-AMP | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, ACETIC ACID, Lipoyltransferase 1, ... | | Authors: | Fujiwara, K, Hosaka, H, Matsuda, M, Suzuki, M, Nakagawa, A. | | Deposit date: | 2006-12-19 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bovine Lipoyltransferase in complex with lipoyl-AMP
J.Mol.Biol., 371, 2007
|
|
2RIF
 
 | | CBS domain protein PAE2072 from Pyrobaculum aerophilum complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CESIUM ION, Conserved protein with 2 CBS domains | | Authors: | Lee, T.M, King, N.P, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures and Functional Implications of an AMP-Binding Cystathionine beta-Synthase Domain Protein from a Hyperthermophilic Archaeon.
J.Mol.Biol., 380, 2008
|
|
5F74
 
 | | Crystal structure of ChREBP:14-3-3 complex bound with AMP | | Descriptor: | 14-3-3 protein beta/alpha, ADENOSINE MONOPHOSPHATE, Carbohydrate-responsive element-binding protein | | Authors: | Jung, H, Uyeda, K. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Metabolite Regulation of Nuclear Localization of Carbohydrate-response Element-binding Protein (ChREBP): ROLE OF AMP AS AN ALLOSTERIC INHIBITOR.
J.Biol.Chem., 291, 2016
|
|
3QOP
 
 | |
5W10
 
 | | Lcd1 GAF domain in complex with cAMP ligand | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cGMP-specific phosphodiesterase | | Authors: | Guzzo, C.R, Maciel, N.K, Barbosa, A.S, Farah, C.S. | | Deposit date: | 2017-06-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Enzymatic Characterization of a cAMP-Dependent Diguanylate Cyclase from Pathogenic Leptospira Species.
J. Mol. Biol., 429, 2017
|
|
1FBP
 
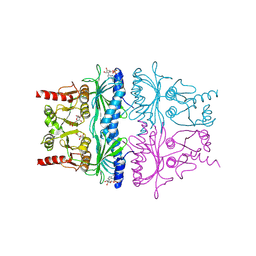 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE 6-PHOSPHATE, AMP, AND MAGNESIUM | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Ke, H, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1990-05-31 | | Release date: | 1992-04-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 6-phosphate, AMP, and magnesium.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
4AVB
 
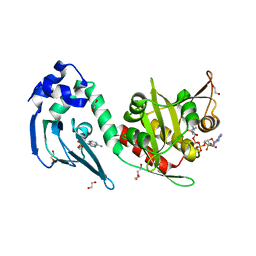 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1G6N
 
 | | 2.1 ANGSTROM STRUCTURE OF CAP-CAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | Passner, J.M, Schultz, S.C, Steitz, T.A. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution.
J.Mol.Biol., 304, 2000
|
|
4AVC
 
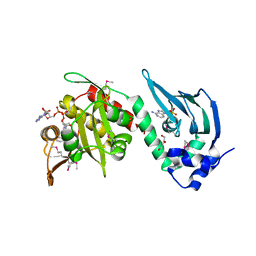 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6AEL
 
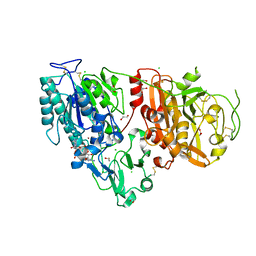 | | Crystal structure of ENPP1 in complex with 3'3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
2GM3
 
 | | Crystal Structure of an Universal Stress Protein Family Protein from Arabidopsis Thaliana At3g01520 with AMP Bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, unknown protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Crystal structure of the protein At3g01520, a eukaryotic universal stress protein-like protein from arabidopsis thaliana in complex with AMP.
Proteins, 83, 2015
|
|
9NTF
 
 | | Helix pomatia AMP deaminase (HPAMPD) with unknown density in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 301, 2025
|
|
9NTE
 
 | | Helix pomatia AMP deaminase (HPAMPD) apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 301, 2025
|
|
7AER
 
 | | Rebuilt and re-refined PDB entry 5yep: tri-AMPylated Shewanella oneidensis HEPN toxin in complex with MNT antitoxin | | Descriptor: | ADENOSINE MONOPHOSPHATE, Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
2A00
 
 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
4IAC
 
 | |
6AIC
 
 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2A29
 
 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
2H6D
 
 | | Protein Kinase Domain of the Human 5'-AMP-activated protein kinase catalytic subunit alpha-2 (AMPK alpha-2 chain) | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Littler, D.R, Walker, J.R, Wybenga-Groot, L, Newman, E.M, Butler-Cole, C, Mackenzie, F, Finerty, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-31 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A conserved mechanism of autoinhibition for the AMPK kinase domain: ATP-binding site and catalytic loop refolding as a means of regulation.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3I54
 
 | | Crystal structure of MtbCRP in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Transcriptional regulator, Crp/Fnr family | | Authors: | Reddy, M.C, Palaninathan, S.K, Bruning, J.B, Thurman, C, Smith, D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Mechanism of the Allosteric Transitions of Mycobacterium tuberculosis cAMP Receptor Protein.
J.Biol.Chem., 284, 2009
|
|
4DFX
 
 | | Crystal structure of myristoylated K7C catalytic subunit of cAMP-dependent protein kinase in complex with SP20 and AMP-PNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, MYRISTIC ACID, ... | | Authors: | Bastidas, A.C, Steichen, J.M, Taylor, S.S. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Role of N-terminal myristylation in the structure and regulation of cAMP-dependent protein kinase.
J.Mol.Biol., 422, 2012
|
|
