4EDM
 
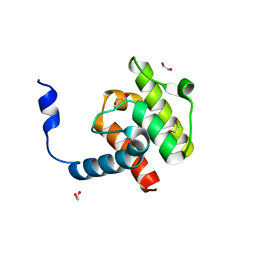 | | Crystal structure of beta-parvin CH2 domain | | Descriptor: | 1,2-ETHANEDIOL, Beta-parvin | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
2Q5E
 
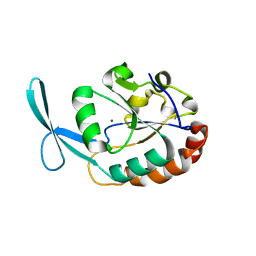 | | Crystal structure of human carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2 | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
8XFU
 
 | |
2Q7T
 
 | |
7UGA
 
 | | Solution structure of NPSL2 | | Descriptor: | NPSL2 RNA (43-MER) | | Authors: | Liu, Y, Keane, S.C. | | Deposit date: | 2022-03-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of NPSL2, A Regulatory Element in the oncomiR-1 RNA.
J.Mol.Biol., 434, 2022
|
|
7OPX
 
 | | CryoEM structure of human enterovirus 70 native virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Fuzik, T, Plevka, P, Moravcova, J. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
J.Virol., 96, 2022
|
|
2QEJ
 
 | | Crystal structure of a Staphylococcus aureus protein (SSL7) in complex with Fc of human IgA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Ramsland, P.A, Willoughby, N, Trist, H.M, Farrugia, W, Hogarth, P.M, Fraser, J.D, Wines, B.D. | | Deposit date: | 2007-06-26 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QOE
 
 | | Human Dipeptidyl Peptidase IV in complex with a Triazolopiperazine-based beta amino acid Inhibitor | | Descriptor: | (2R)-4-[(8R)-8-METHYL-2-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[1,5-A]PYRAZIN-7(8H)-YL]-4-OXO-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-AMINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2007-07-20 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological evaluation of triazolopiperazine-based beta-amino amides as potent, orally active dipeptidyl peptidase IV (DPP-4) inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8GZ1
 
 | | Cryo-EM structure of human NaV1.6/beta1/beta2,apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
8GZ2
 
 | | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | | Descriptor: | (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
2QCU
 
 | | Crystal structure of Glycerol-3-phosphate Dehydrogenase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Aerobic glycerol-3-phosphate dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yeh, J.I, Chinte, U, Du, S. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7V33
 
 | | Active state complex I from rotenone-NADH dataset | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2D
 
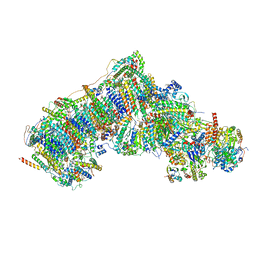 | | Deactive state complex I from Q10 dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-08 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2R
 
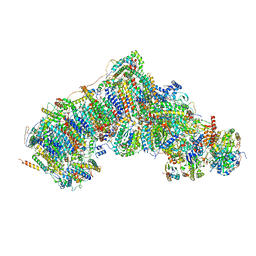 | | Active state complex I from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2K
 
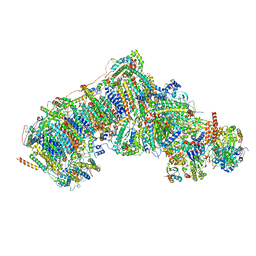 | | Deactive state complex I from DQ-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2F
 
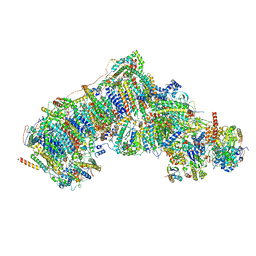 | | Deactive state complex I from Q10-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4G9J
 
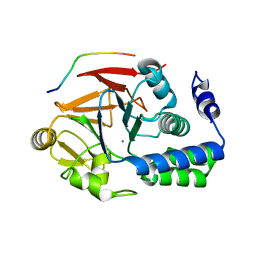 | | Protein Ser/Thr phosphatase-1 in complex with cell-permeable peptide | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, synthetic peptide | | Authors: | Sukackaite, R, Chatterjee, J, Beullens, M, Bollen, M, Koehn, M, Hart, D.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of a Peptide that Selectively Activates Protein Phosphatase-1 in Living Cells.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
8HCD
 
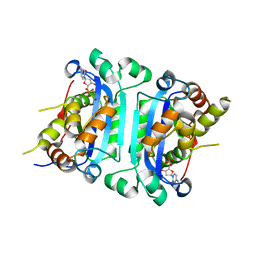 | | Crystal structure of mTREX1-DNA product complex (dNMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, COBALT (II) ION, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCG
 
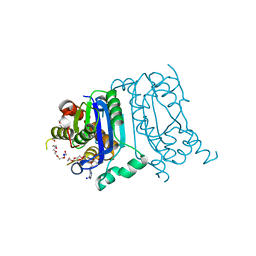 | | Crystal structure of mTREX1-dAMP complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCE
 
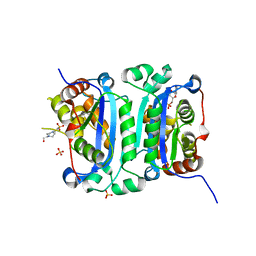 | | Crystal structure of mTREX1-CMP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCC
 
 | | Crystal structure of mTREX1-RNA product complex (AMP) | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCF
 
 | | Crystal structure of mTREX1-UMP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Three-prime repair exonuclease 1, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCH
 
 | | Crystal structure of mTREX1-Uridine complex | | Descriptor: | MAGNESIUM ION, Three-prime repair exonuclease 1, URIDINE | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
7V59
 
 | | Cryo-EM structure of spyCas9-sgRNA-DNA dimer | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (49-MER), RNA (115-MER) | | Authors: | Liu, J, Deng, P. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.26 Å) | | Cite: | Nonspecific interactions between SpCas9 and dsDNA sites located downstream of the PAM mediate facilitated diffusion to accelerate target search.
Chem Sci, 12, 2021
|
|
4DSA
 
 | | Crystal Structure of DPP-IV with Compound C1 | | Descriptor: | 4-[({[(2R)-2-amino-3-(2,4,5-trifluorophenyl)propyl]sulfamoyl}amino)methyl]benzenesulfonamide, Dipeptidyl peptidase 4 | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2012-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structure of DPP-IV with Compound C1
To be Published
|
|
