1B8X
 
 | | GLUTATHIONE S-TRANSFERASE FUSED WITH THE NUCLEAR MATRIX TARGETING SIGNAL OF THE TRANSCRIPTION FACTOR AML-1 | | Descriptor: | PROTEIN (AML-1B) | | Authors: | Tang, L, Guo, B, Van Wijnen, A.J, Lian, J.B, Stein, J.L, Stein, G.S, Zhou, G.W. | | Deposit date: | 1999-02-03 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preliminary crystallographic study of glutathione S-transferase fused with the nuclear matrix targeting signal of the transcription factor AML-1/CBF-alpha2.
J.Struct.Biol., 123, 1998
|
|
1IPK
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS | | Descriptor: | BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
4HFW
 
 | | Anti Rotavirus Antibody | | Descriptor: | 6-26 Fab Heavy chain, 6-26 Fab Light chain, SULFATE ION | | Authors: | Spiller, B.W, Aiyegbo, M, Crowe, J.E. | | Deposit date: | 2012-10-05 | | Release date: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Human Rotavirus VP6-Specific Antibodies Mediate Intracellular Neutralization by Binding to a Quaternary Structure in the Transcriptional Pore.
Plos One, 8, 2013
|
|
4HIL
 
 | | 1.25A Resolution Structure of Rat Type B Cytochrome b5 | | Descriptor: | Cytochrome b5 type B, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25A Resolution Structure of Rat Type B Cytochrome b5
To be Published
|
|
1IN3
 
 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
2MQK
 
 | |
1BBR
 
 | |
1BTX
 
 | |
1JC2
 
 | | COMPLEX OF THE C-DOMAIN OF TROPONIN C WITH RESIDUES 1-40 OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, SKELETAL MUSCLE | | Authors: | Mercier, P, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 2001-06-07 | | Release date: | 2001-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and thermodynamics of the structural domain of troponin C in complex with the regulatory peptide 1-40 of troponin I
Biochemistry, 40, 2001
|
|
3FAW
 
 | | Crystal Structure of the Group B Streptococcus Pullulanase SAP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Reticulocyte binding protein | | Authors: | Gourlay, L.J. | | Deposit date: | 2008-11-18 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group B Streptococcus pullulanase crystal structures in the context of a novel strategy for vaccine development
J.Bacteriol., 191, 2009
|
|
3K9Z
 
 | | Rational Design of a Structural and Functional Nitric Oxide Reductase | | Descriptor: | FE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yeung, N, Lin, Y.-W, Gao, Y.-G, Zhao, X, Russell, B.S, Lei, L, Miner, K.D, Robinson, H, Lu, Y. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of a structural and functional nitric oxide reductase.
Nature, 462, 2009
|
|
1BTZ
 
 | |
3ZHF
 
 | | gamma 2 adaptin EAR domain crystal structure with preS1 site1 peptide NPDWDFN | | Descriptor: | 1,2-ETHANEDIOL, AP-1 COMPLEX SUBUNIT GAMMA-LIKE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Juergens, M.C, Voros, J, Rautureau, G, Shepherd, D, Pye, V.E, Muldoon, J, Johnson, C.M, Ashcroft, A, Freund, S.M.V, Ferguson, N. | | Deposit date: | 2012-12-21 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Hepatitis B Virus Pres1 Domain Hijacks Host Trafficking Proteins by Motif Mimicry.
Nat.Chem.Biol., 9, 2013
|
|
1WB8
 
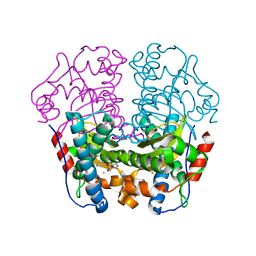 | | Iron Superoxide Dismutase (FE-SOD) from the Hyperthermophile SULFOLOBUS SOLFATARICUS. 2.3 A Resolution Structure of Recombinant Protein with a Covalently Modified Tyrosine in the Active Site. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE [FE], phenylmethanesulfonic acid | | Authors: | Ursby, T, Adinolfi, B.S, Al-Karadaghi, S, De Vendittis, E, Bocchini, V. | | Deposit date: | 2004-10-31 | | Release date: | 2004-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Iron Superoxide Dismutase from the Archaeon Sulfolobus Solfataricus: Analysis of Structure and Thermostability
J.Mol.Biol., 286, 1999
|
|
1K3M
 
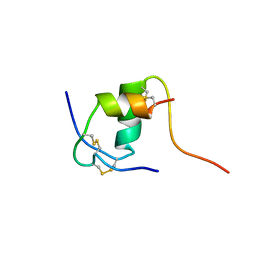 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALA, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Xu, B, Hua, Q.-X, Nakagawa, S.H, Jia, W, Chu, Y.-C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-17 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A cavity-forming mutation in insulin induces segmental unfolding of a surrounding alpha-helix.
Protein Sci., 11, 2002
|
|
1YI2
 
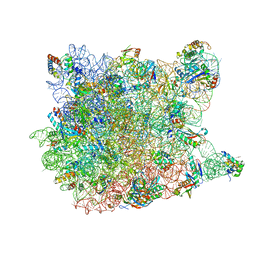 | | Crystal Structure Of Erythromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-11 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1W6G
 
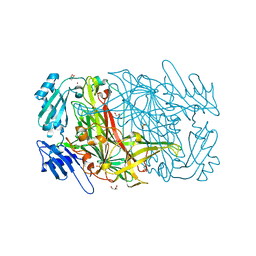 | | AGAO holoenzyme at 1.55 angstroms | | Descriptor: | COPPER (II) ION, GLYCEROL, PHENYLETHYLAMINE OXIDASE, ... | | Authors: | Langley, D.B, Duff, A.P, Juda, G.A, Shepard, E.M, Dooley, D.M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-08-18 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Copper Containing Amine Oxidase from Arthrobacter Globiformis: Refinement at 1.55 And 2.20 A Resolution in Two Crystal Forms.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
4FLV
 
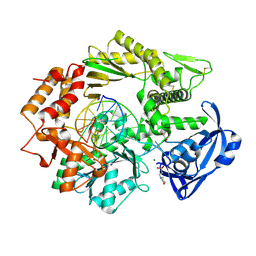 | |
1W52
 
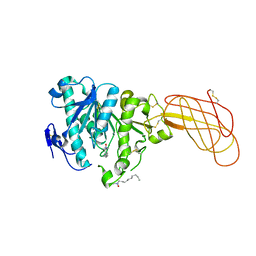 | | Crystal structure of a proteolyzed form of pancreatic lipase related protein 2 from horse | | Descriptor: | CALCIUM ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, PANCREATIC LIPASE RELATED PROTEIN 2 | | Authors: | Mancheno, J.M, Jayne, S, Kerfelec, B, Chapus, C, Crenon, I, Hermoso, J.A. | | Deposit date: | 2004-08-04 | | Release date: | 2006-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystalization of a Proteolyzed Form of the Horse Pancreatic Lipase-Related Protein 2: Structural Basis for the Specific Detergent Requirement.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4FKA
 
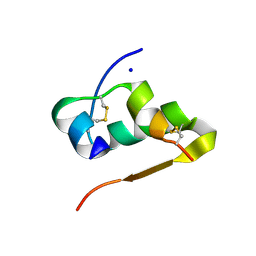 | | High resolution structure of the manganese derivative of insulin | | Descriptor: | Insulin A chain, Insulin B chain, MANGANESE (II) ION, ... | | Authors: | Prugovecki, B, Pulic, I, Toth, M, Matkovic-Calogovic, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High Resolution Structure of the Manganese Derivative of Insulin
Croat.Chem.Acta, 85, 2012
|
|
4FM0
 
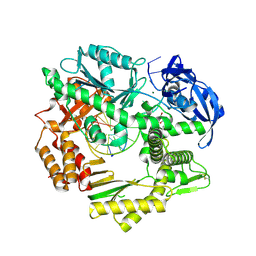 | |
1CBU
 
 | | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE (COBU) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE, SULFATE ION | | Authors: | Thompson, T.B, Thomas, M.G, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 1998-03-12 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase from Salmonella typhimurium determined to 2.3 A resolution,.
Biochemistry, 37, 1998
|
|
1C8K
 
 | | FLAVOPIRIDOL INHIBITS GLYCOGEN PHOSPHORYLASE BY BINDING AT THE INHIBITOR SITE | | Descriptor: | 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site.
J.Biol.Chem., 275, 2000
|
|
4FRQ
 
 | | Crystal Structure of BBBB+UDP+Gal at pH 9.5 with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
2BX9
 
 | | Crystal structure of B.subtilis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | TRYPTOPHAN RNA-BINDING ATTENUATOR PROTEIN-INHIBITORY PROTEIN, ZINC ION | | Authors: | Shevtsov, M.B, Chen, Y, Gollnick, P, Antson, A.A. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Anti-Trap Protein, an Antagonist of Trap/RNA Interaction.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
