7CP4
 
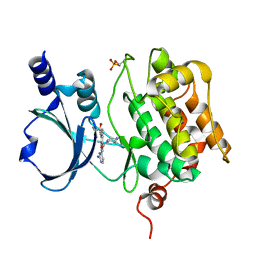 | | Crystal Structure of PAK4 in complex with inhibitor 55 | | Descriptor: | Serine/threonine-protein kinase PAK 4, [1-(2-azanylpyrimidin-4-yl)-6-[2-(1-oxidanylcyclohexyl)ethynyl]indol-3-yl]-[(3S)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2020-08-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of 6-ethynyl-1H-indole-3-carboxamide Derivatives as Highly Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors
to be published
|
|
4ETU
 
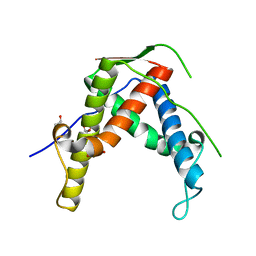 | |
7GYB
 
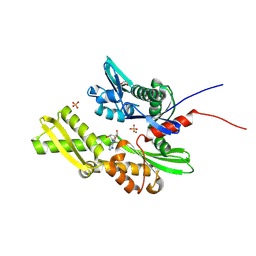 | | Crystal Structure of HSP72 in complex with ligand 10 at 9.12 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
6ITT
 
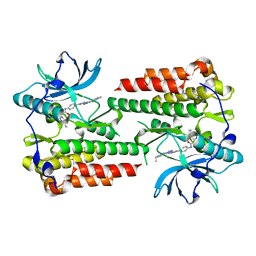 | | Crystal structure of unactivated c-KIT in complex with compound | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-(5-ethyl-1,2-oxazol-3-yl)-N'-[4-(2-{[6-(4-ethylpyrazin-1(4H)-yl)pyrimidin-4-yl]amino}-1,3-thiazol-5-yl)phenyl]urea | | Authors: | Wu, T.S, Wu, S.Y. | | Deposit date: | 2018-11-26 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of unactivated c-KIT in complex with compound
To Be Published
|
|
5L15
 
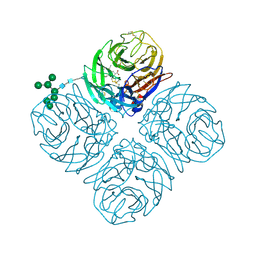 | | The crystal structure of neuraminidase in complex with oseltamivir from A/Shanghai/2/2013 (H7N9) influenza virus | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Stevens, J. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Drug Susceptibility Evaluation of an Influenza A(H7N9) Virus by Analyzing Recombinant Neuraminidase Proteins.
J. Infect. Dis., 216, 2017
|
|
7GYD
 
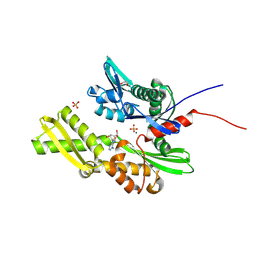 | | Crystal Structure of HSP72 in complex with ligand 10 at 11.40 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
4QMG
 
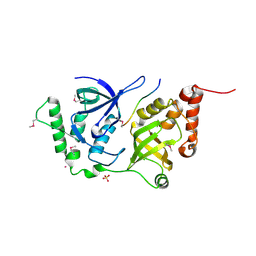 | | The Structure of MTDH-SND1 Complex Reveals Novel Cancer-Promoting Interactions | | Descriptor: | CESIUM ION, GLYCEROL, Protein LYRIC, ... | | Authors: | Guo, F, Stanevich, V, Wan, L, Satyshur, K, Kang, Y, Xing, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural Insights into the Tumor-Promoting Function of the MTDH-SND1 Complex.
Cell Rep, 8, 2014
|
|
7GYE
 
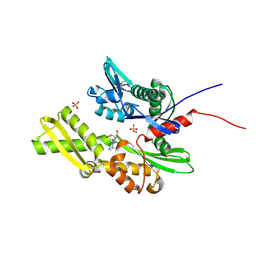 | | Crystal Structure of HSP72 in complex with ligand 10 at 12.54 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
3LLR
 
 | | Crystal structure of the PWWP domain of Human DNA (cytosine-5-)-methyltransferase 3 alpha | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (cytosine-5)-methyltransferase 3A, SULFATE ION | | Authors: | Qiu, W, Dombrovski, L, Ni, S, Weigelt, J, Boutra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
8QTW
 
 | | Structure of human butyrylcholinesterase with (3-(((2-cycloheptylethyl)(methyl)amino)methyl)-1H-indol-7-yl)(methyl)carbamoylated Ser198 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Meden, A, Knez, D, Gobec, S, Nachon, F. | | Deposit date: | 2023-10-13 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of human butyrylcholinesterase with (3-(((2-cycloheptylethyl)(methyl)amino)methyl)-1H-indol-7-yl)(methyl)carbamoylated Ser198
To Be Published
|
|
6CX7
 
 | | Structure of alpha-GSA[12,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
7GY6
 
 | | Crystal Structure of HSP72 in complex with ligand 10 at 3.42 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GY9
 
 | | Crystal Structure of HSP72 in complex with ligand 10 at 6.84 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
5SLL
 
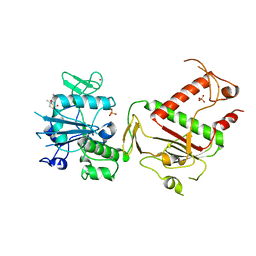 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z54615640 | | Descriptor: | N-[(3R)-3-methyl-1,1-dioxo-1lambda~6~-thiolan-3-yl]cyclopropanecarboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLZ
 
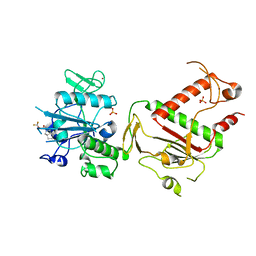 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2072621991 | | Descriptor: | 2-(difluoromethoxy)-1-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-yl]ethan-1-one, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4QLU
 
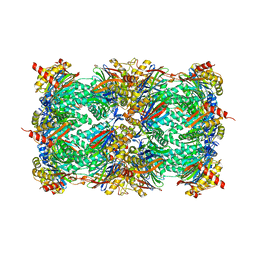 | | yCP in complex with tripeptidic epoxyketone inhibitor 9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(3-methyl-1H-inden-2-yl)carbonyl]-D-alanyl-N-[(2S,4R)-1-cyclohexyl-5-hydroxy-4-methyl-3-oxopentan-2-yl]-L-tryptophanamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4Q5J
 
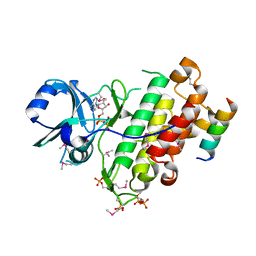 | | Crystal structure of SeMet derivative BRI1 in complex with BKI1 | | Descriptor: | BRI1 kinase inhibitor 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein BRASSINOSTEROID INSENSITIVE 1 | | Authors: | Wang, J, Wang, J, Chen, L, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-04-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.772 Å) | | Cite: | Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1.
Cell Res., 24, 2014
|
|
1IS5
 
 | | Ligand free Congerin II | | Descriptor: | Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
5SMF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z56791867 | | Descriptor: | N,N-diethyl-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2DRR
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263N mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
251L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
5EVK
 
 | | Crystal structure of the metallo-beta-lactamase L1 in complex with the bisthiazolidine inhibitor L-CS319 | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.627 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5PB2
 
 | | Crystal Structure of Factor VIIa in complex with 2-phenyl-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-3-ol | | Descriptor: | 1-phenyl-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)-1H-pyrazol-5-ol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
4EJ5
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A wild-type | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, CARBONATE ION, ... | | Authors: | Stura, E.A, Vera, L, Dive, V. | | Deposit date: | 2012-04-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
5EYR
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 5 at 1.57A resolution | | Descriptor: | 3-[1-[4-(methylaminomethyl)phenyl]piperidin-4-yl]-1-pyrrolidin-1-yl-propan-1-one, EthR | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-25 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
