8UN4
 
 | |
8QV0
 
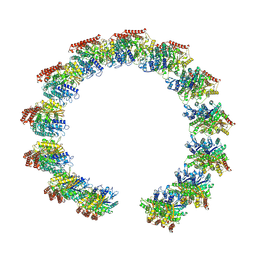 | | Structure of the native microtubule lattice nucleated from the yeast spindle pole body | | Descriptor: | Tubulin alpha-1 chain, Tubulin beta chain | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 2024
|
|
8QV3
 
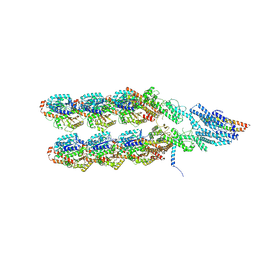 | | Structure of the y-Tubulin Small Complex (yTuSC) as part of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component, Spindle pole body component 110, ... | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 2024
|
|
8QV2
 
 | | Structure of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component, ... | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 2024
|
|
8WSS
 
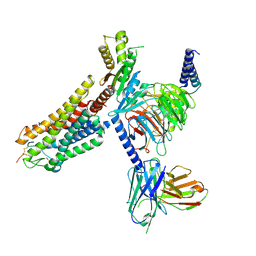 | | Cryo-EM structure of Melanin-Concentrating Hormone Receptor 1 with MCH | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, Shan, H, Hu, W, Wu, K, Xu, H.E, Zhang, Y, Wu, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Mechanisms of ligand recognition and activation of melanin-concentrating hormone receptors.
Cell Discov, 10, 2024
|
|
8WST
 
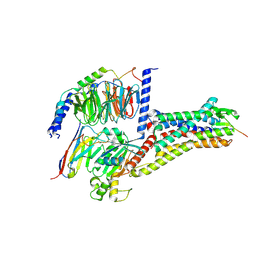 | | Cryo-EM structure of Melanin-Concentrating Hormone Receptor 2 with MCH | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, Shan, H, Hu, W, Wu, K, Xu, H.E, Zhang, Y, Wu, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanisms of ligand recognition and activation of melanin-concentrating hormone receptors.
Cell Discov, 10, 2024
|
|
8WRZ
 
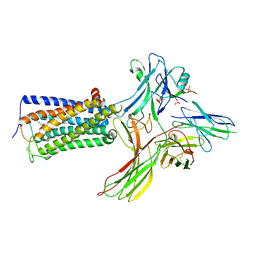 | | Cry-EM structure of cannabinoid receptor-arrestin 2 complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Beta-arrestin-1, Soluble cytochrome b562,Cannabinoid receptor 1, ... | | Authors: | Wang, Y.X, Wang, T, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of cannabinoid receptor CB1-beta-arrestin complex.
Protein Cell, 15, 2024
|
|
8QUG
 
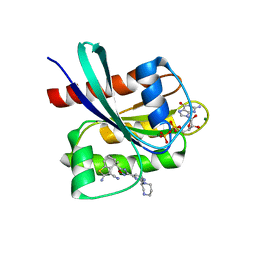 | | KRAS-G12C in Complex with Compound 1 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[2-[(2S)-2-methyl-1,4-diazepan-1-yl]pyrimidin-4-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fischer, G, Kratochvil, B. | | Deposit date: | 2023-10-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Targeting cancer with small molecule pan-KRAS degraders
Biorxiv, 2023
|
|
8UKZ
 
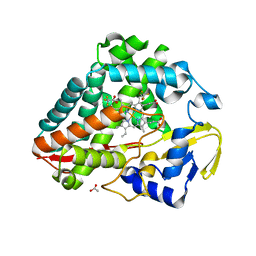 | |
8QU8
 
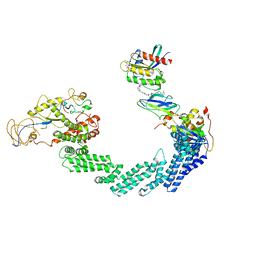 | | PROTAC-mediated complex of KRAS with VHL/Elongin-B/Elongin-C/Cullin-2/Rbx1 | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Fischer, G, Peter, D, Arce-Solano, S. | | Deposit date: | 2023-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting cancer with small molecule pan-KRAS degraders
Biorxiv, 2023
|
|
8WRB
 
 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
8QTO
 
 | | CRYSTAL STRUCTURE OF HOLO-L28H-FNR OF A. FISCHERI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FNR type regulator, IRON/SULFUR CLUSTER | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Reactivity of [4Fe-4S] Fumarate and Nitrate Reduction (FNR) Regulator with O2 and NO: Increased O2 Resistance and Relative Specificity for NO of the [4Fe-4S] L28H FNR Cluster
Inorganics (Basel), 11, 2023
|
|
8UK9
 
 | | Structure of T4 Bacteriophage clamp loader mutant D110C bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA primer, ... | | Authors: | Marcus, K, Ghaffari-Kashani, S, Gee, C.L. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QTG
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 9) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-5-(trifluoromethyl)-1~{H}-pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTH
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 8) | | Descriptor: | 1-methyl-5-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-(trifluoromethyl)-7H-pyrrolo[2,3-b]pyridin-6-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTJ
 
 | | Crystal structure of Cbl-b in complex with an allosteric inhibitor (compound 30) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-1-[(1~{R})-1-(1-methylpyrazol-4-yl)ethyl]-5-(trifluoromethyl)pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QTK
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 31) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-1-[(1S)-1-(1-methylpyrazol-4-yl)ethyl]-5-(trifluoromethyl)pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8WPU
 
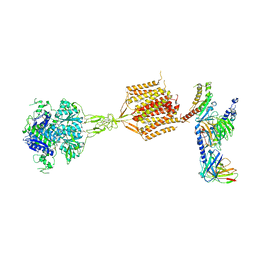 | | Human calcium-sensing receptor(CaSR) bound to cinacalcet in complex with Gq protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
8WPG
 
 | | Human calcium-sensing receptor bound with cinacalcet in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
8WPZ
 
 | |
8UI8
 
 | |
8UIN
 
 | | Structure of the C3bBb-albicin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Albicin, ... | | Authors: | Andersen, J.F, Lei, H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Mechanism of complement inhibition by a mosquito protein revealed through cryo-EM.
Commun Biol, 7, 2024
|
|
8UI9
 
 | |
8UI7
 
 | |
8UII
 
 | |
