3MFI
 
 | | DNA Polymerase Eta in Complex With a cis-syn Thymidine Dimer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*TP*CP*CP*TP*CP*CP*CP*CP*TP*(DOC))-3', 5'-D(*TP*AP*AP*(TTD)P*GP*AP*GP*GP*GP*GP*AP*GP*GP*AP*C)-3', ... | | Authors: | Silverstein, T.D, Johnson, R.E, Jain, R, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2010-04-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the suppression of skin cancers by DNA polymerase eta.
Nature, 465, 2010
|
|
1ZQZ
 
 | |
1ZQY
 
 | |
1ZQX
 
 | |
1ZQW
 
 | |
1ZQU
 
 | |
1BP7
 
 | | GROUP I MOBILE INTRON ENDONUCLEASE I-CREI COMPLEXED WITH HOMING SITE DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP* GP*C)-3'), DNA (5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP* CP*G)-3'), ... | | Authors: | Jurica, M.S, Monnat Junior, R.J, Stoddard, B.L. | | Deposit date: | 1998-08-13 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI.
Mol.Cell, 2, 1998
|
|
1KLN
 
 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*CP*GP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*CP*GP*CP*GP*GP*CP*GP*GP*C)-3'), PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Beese, L.S, Derbyshire, V, Steitz, T.A. | | Deposit date: | 1994-05-24 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of DNA polymerase I Klenow fragment bound to duplex DNA.
Science, 260, 1993
|
|
3D1G
 
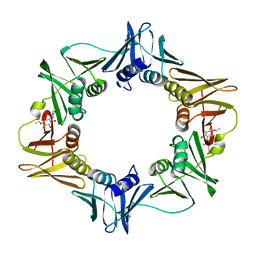 | | Structure of a small molecule inhibitor bound to a DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1UW0
 
 | |
6MHT
 
 | |
5YUY
 
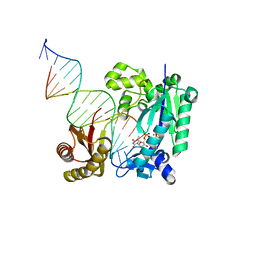 | | DNA polymerase IV - DNA ternary complex 9 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Nucleic Acids Res., 46, 2018
|
|
5YUZ
 
 | | DNA polymerase IV - DNA ternary complex 11 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN1, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction.
Nucleic Acids Res., 46, 2018
|
|
5YUX
 
 | | DNA polymerase IV - DNA ternary complex 8 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN1, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Nucleic Acids Res., 46, 2018
|
|
5YYD
 
 | | DNA polymerase IV - ternary complex 15 | | Descriptor: | 5'-O-[hydroxy{[hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]thymidine, DNA polymerase IV, DTN2, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-12-08 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Nucleic Acids Res., 46, 2018
|
|
5YV3
 
 | | DNA polymerase IV - DNA ternary complex 7 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN1, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Nucleic Acids Res., 46, 2018
|
|
5YV0
 
 | | DNA polymerase IV - DNA ternary complex 12 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN1, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction.
Nucleic Acids Res., 46, 2018
|
|
2ZHX
 
 | | Crystal structure of Uracil-DNA Glycosylase from Mycobacterium tuberculosis in complex with a proteinaceous inhibitor | | Descriptor: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Kaushal, P.S, Talawar, R.K, Krishna, P.D.V, Varshney, U, Vijayan, M. | | Deposit date: | 2008-02-11 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unique features of the structure and interactions of mycobacterial uracil-DNA glycosylase: structure of a complex of the Mycobacterium tuberculosis enzyme in comparison with those from other sources
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3FL6
 
 | |
5ZLV
 
 | |
5YYE
 
 | | DNA polymerase IV - ternary complex 16 | | Descriptor: | 5'-O-[hydroxy{[hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]thymidine, DNA polymerase IV, DTN2, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-12-08 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.325 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Nucleic Acids Res., 46, 2018
|
|
5YV1
 
 | | DNA polymerase IV - DNA ternary complex 13 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN1, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Nucleic Acids Res., 46, 2018
|
|
1KFS
 
 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*AP*CP*G)-3'), MAGNESIUM ION, PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
2BDP
 
 | | CRYSTAL STRUCTURE OF BACILLUS DNA POLYMERASE I FRAGMENT COMPLEXED TO 9 BASE PAIRS OF DUPLEX DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*AP*GP*CP*AP*TP*CP*AP*TP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Kiefer, J.R, Mao, C, Beese, L.S. | | Deposit date: | 1997-11-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal.
Nature, 391, 1998
|
|
6IG1
 
 | | DNA polymerase IV - DNA ternary complex 10 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN3, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2018-09-21 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction.
Nucleic Acids Res., 46, 2018
|
|
