3IXF
 
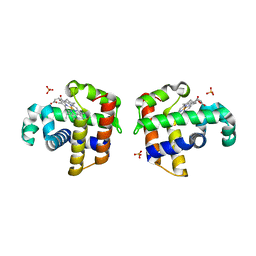 | | Crystal Structure of Dehaloperoxidase B at 1.58 and Structural Characterization of the AB Dimer from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Serrano, V.S, D'Antonio, J, Thompson, M.K, Franzen, S, Ghiladi, R.A. | | Deposit date: | 2009-09-03 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of dehaloperoxidase B at 1.58 A resolution and structural characterization of the AB dimer from Amphitrite ornata.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4LE5
 
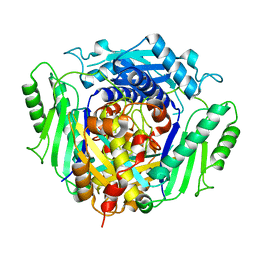 | |
5LDT
 
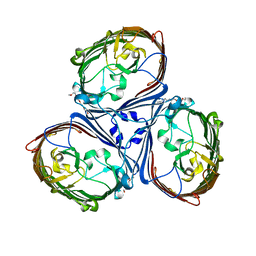 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, MOMP porin | | Authors: | Ferrara, L.G.M, Wallat, G.D, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
4MT4
 
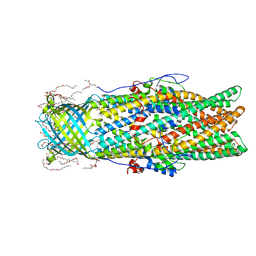 | |
4GIO
 
 | | Crystal structure of Campylobacter jejuni cj0090 | | Descriptor: | BROMIDE ION, Putative lipoprotein | | Authors: | Kawai, F, Yeo, H.J. | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Campylobacter jejuni Cj0090 protein reveals a novel variant of the immunoglobulin fold among bacterial lipoproteins.
Proteins, 80, 2012
|
|
3HBM
 
 | | Crystal Structure of PseG from Campylobacter jejuni | | Descriptor: | SULFATE ION, UDP-sugar hydrolase | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
3QPS
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QQA
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
4BOL
 
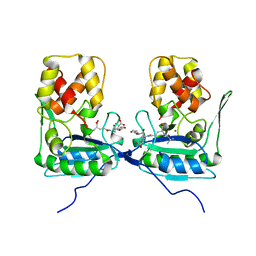 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with pentapeptide | | Descriptor: | AMPDH2, D-alanyl-N-[(2S,6R)-6-amino-6-carboxy-1-{[(1R)-1-carboxyethyl]amino}-1-oxohexan-2-yl]-D-glutamine, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E.E, Lastochkin, E, Zhang, W, Hellman, L.M, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
3KUN
 
 | | X-ray structure of the metcyano form of dehaloperoxidase from amphitrite ornata: evidence for photoreductive lysis of iron-cyanide bond | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Serrano, V.S, Chen, Z, Gaff, J.F, Rose, R, Franzen, S. | | Deposit date: | 2009-11-27 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | X-ray structure of the metcyano form of dehaloperoxidase from Amphitrite ornata: evidence for photoreductive dissociation of the iron-cyanide bond.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1RO7
 
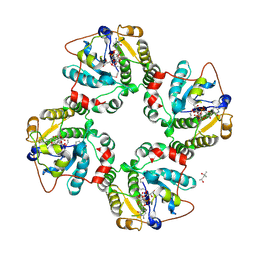 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, CMP-3FNeuAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RO8
 
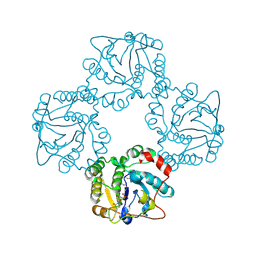 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
8IVU
 
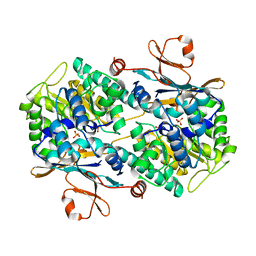 | | Crystal Structure of Human NAMPT in complex with A4276 | | Descriptor: | N-[[4-(6-methyl-1,3-benzoxazol-2-yl)phenyl]methyl]pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Kang, B.G, Cha, S.S. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09000921 Å) | | Cite: | Discovery of a novel NAMPT inhibitor that selectively targets NAPRT-deficient EMT-subtype cancer cells and alleviates chemotherapy-induced peripheral neuropathy.
Theranostics, 13, 2023
|
|
3QQT
 
 | |
4ILZ
 
 | | Structural and kinetic study of an internal substrate binding site of dehaloperoxidase-hemoglobin A from Amphitrite ornata | | Descriptor: | 2,4,6-TRIBROMOPHENOL, Dehaloperoxidase A, ISOPROPYL ALCOHOL, ... | | Authors: | de Serrano, V.S, Zhao, J, Zhao, J, Franzen, S. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural and Kinetic Study of an Internal Substrate Binding Site in Dehaloperoxidase-Hemoglobin A from Amphitrite ornata.
Biochemistry, 52, 2013
|
|
4KJT
 
 | | Structure of the L100F MUTANT OF DEHALOPEROXIDASE-HEMOGLOBIN A FROM AMPHITRITE ORNATA WITH OXYGEN | | Descriptor: | 1,2-ETHANEDIOL, Dehaloperoxidase A, OXYGEN MOLECULE, ... | | Authors: | Wang, C, Lovelace, L, Lebioda, L. | | Deposit date: | 2013-05-03 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Influence of heme environment structure on dioxygen affinity for the dual function Amphitrite ornata hemoglobin/dehaloperoxidase. Insights into the evolutional structure-function adaptations.
Arch.Biochem.Biophys., 545, 2014
|
|
5TPV
 
 | | X-ray structure of WlaRA (TDP-fucose-3,4-ketoisomerase) from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, THYMIDINE-5'-DIPHOSPHATE, WlaRA, ... | | Authors: | Holden, H.M, Thoden, J.B, Li, Z.A, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinograd, E, Schoenhofen, I.C, Gilbert, M, Li, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
2XF4
 
 | | Crystal structure of Salmonella enterica serovar typhimurium YcbL | | Descriptor: | HYDROXYACYLGLUTATHIONE HYDROLASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Stamp, A, Owen, P, El Omari, K, Nichols, C, Lockyer, M, Lamb, H, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2010-05-20 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Salmonella Enterica Serovar Typhimurium Ycbl: An Unusual Type II Glyoxalase
Protein Sci., 19, 2010
|
|
5TPU
 
 | | x-ray structure of the WlaRB TDP-quinovose 3,4-ketoisomerase from campylobacter jejuni | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Holden, H.M, Thoden, J.B, Li, J.Z, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinogradov, E, Schoenhofen, I.C, Gilbert, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
5TYH
 
 | |
7ENQ
 
 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
4JKN
 
 | | Mercury Metallated Pseudomonas aeruginosa Azurin at 1.54 A | | Descriptor: | Azurin, MERCURY (II) ION, NITRATE ION, ... | | Authors: | Zampino, A.P, Masters, F.M, Bladholm, E.L, Berry, S.B, Panzner, M.J, Ziegler, C.J. | | Deposit date: | 2013-03-10 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.536 Å) | | Cite: | Mercury metallation of the copper protein azurin and structural insight into possible heavy metal reactivity.
J.Inorg.Biochem., 141, 2014
|
|
4ORH
 
 | | Crystal structure of RNF8 bound to the UBC13/MMS2 heterodimer | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.802 Å) | | Cite: | Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
J.Biol.Chem., 287, 2012
|
|
6DK4
 
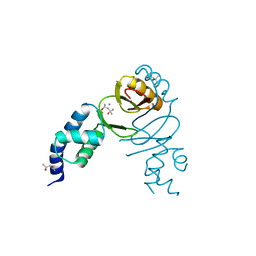 | | Crystal structure of Campylobacter jejuni peroxide stress regulator | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Ferric uptake regulation protein, MANGANESE (II) ION, ... | | Authors: | Sarvan, S, Brunzelle, J.S, Couture, J.F. | | Deposit date: | 2018-05-28 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of Campylobacter jejuni peroxide regulator.
FEBS Lett., 592, 2018
|
|
6CF9
 
 | |
