5Y2Y
 
 | | Crystal structure of HaloTag (M175C) complexed with dansyl-PEG2-HaloTag ligand | | Descriptor: | 5-(dimethylamino)-~{N}-[2-(2-hexoxyethoxy)ethyl]naphthalene-1-sulfonamide, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lee, H, Kang, M, Rhee, H, Lee, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-guided synthesis of a protein-based fluorescent sensor for alkyl halides
Chem. Commun. (Camb.), 53, 2017
|
|
6FZR
 
 | | Crystal structure of scFv-SM3 in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-[(fluoroacetyl)amino]-alpha-D-galactopyranose, Mucin-1, ... | | Authors: | Bermejo, I.A, Usabiaga, I, Companon, I, Castro-Lopez, J, Insausti, A, Fernandez, J.A, Avenoza, A, Busto, J.H, Jimenez-Barbero, J, Asensio, J.L, Jimenez-Oses, G, Peregrina, J.M, Hurtado-Guerrero, R, Cocinero, E.J, Corzana, F. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water Sculpts the Distinctive Shapes and Dynamics of the Tumor-Associated Carbohydrate Tn Antigens: Implications for Their Molecular Recognition.
J.Am.Chem.Soc., 140, 2018
|
|
6VDS
 
 | | Crystal Structure of Dehaloperoxidase B in Complex with cofactor Iron(III) Deuteroporphyrin IX and Substrate 4-bromo-ortho-cresol | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-2-methylphenol, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, McGuire, A, Malewschik, T. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
5S6Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with PB2255187532 | | Descriptor: | 4-[(dimethylamino)methyl]-1,3-thiazol-2-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5C0O
 
 | | m1A58 tRNA methyltransferase mutant - Y78A | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, tRNA (adenine(58)-N(1))-methyltransferase TrmI | | Authors: | Degut, C, Ponchon, L, Folly-Klan, M, Barraud, P, Tisne, C. | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The m1A58 modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.
Biophys.Chem., 210, 2016
|
|
5CAO
 
 | | EGFR kinase domain mutant "TMLR" with compound 29 | | Descriptor: | Epidermal growth factor receptor, N~2~-[2-methyl-2-(methylsulfonyl)propyl]-N~4~-[2-methyl-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-yl]pyrimidine-2,4-diamine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
6URX
 
 | | Crystal structure of ricin A chain in complex with inhibitor 5-phenyl-2-thiophenecarboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-phenylthiophene-2-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Harijan, R.K, Li, X.P, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Small Molecule Inhibitors Targeting the Interaction of Ricin Toxin A Subunit with Ribosomes.
Acs Infect Dis., 6, 2020
|
|
7ERX
 
 | | Glycosyltransferase in complex with UDP and STB | | Descriptor: | GLYCEROL, Glycosyltransferase, Steviolbioside, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
5Z4F
 
 | | An anthrahydroquino-Gama-pyrone synthase Txn09 complexed with SUM | | Descriptor: | 1,8-dihydroxy-2-[(4R)-4-hydroxy-4-methyl-3-oxohexanoyl]-3-methylanthracene-9,10-dione, TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4BAR
 
 | | Thaumatin from Thaumatococcus daniellii structure in complex with the europium tris-hydroxyethyltriazoledipicolinate complex at 1.20 A resolution. | | Descriptor: | 4-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, EUROPIUM (III) ION, THAUMATIN-1 | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Clicked europium dipicolinate complexes for protein X-ray structure determination.
Chem. Commun. (Camb.), 48, 2012
|
|
7EPO
 
 | | Ketosteroid Isomerase KSI with 5-nitrobenzoxazole (5NBI) | | Descriptor: | 5-nitro-1,2-benzoxazole, SnoaL-like domain-containing protein | | Authors: | Liang, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-04-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization and Kemp eliminase activity of the Mycobacterium smegmatis Ketosteroid Isomerase.
Biochem.Biophys.Res.Commun., 560, 2021
|
|
5YFB
 
 | | Crystal structure of a new DPP III family member | | Descriptor: | 1,2-ETHANEDIOL, Dipeptidyl peptidase 3, GLYCEROL, ... | | Authors: | Xu, T, Sun, Z, Liu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
5YG0
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS657 | | Descriptor: | 2-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]piperidin-4-yl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | to be published
to be published
|
|
4TTK
 
 | |
7L7L
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-129 | | Descriptor: | 1,1,1-trifluoro-2-methylpropan-2-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
1OJJ
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
4XE3
 
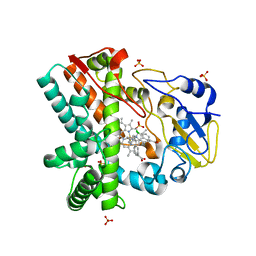 | | OleP, the cytochrome P450 epoxidase from Streptomyces antibioticus involved in Oleandomycin biosynthesis: functional analysis and crystallographic structure in complex with clotrimazole. | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Montemiglio, L.C, Parisi, G, Scaglione, A, Savino, C, Vallone, B. | | Deposit date: | 2014-12-22 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional analysis and crystallographic structure of clotrimazole bound OleP, a cytochrome P450 epoxidase from Streptomyces antibioticus involved in oleandomycin biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
4S18
 
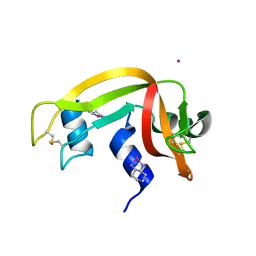 | |
7EXP
 
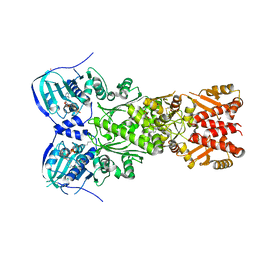 | | Crystal structure of zebrafish TRAP1 with AMPPNP and MitoQ | | Descriptor: | 2,3-dimethoxy-5-methyl-6-[10-(triphenyl-$l^{5}-phosphanyl)decyl]cyclohexa-2,5-diene-1,4-dione, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Lee, H, Yoon, N.G, Kang, B.H, Lee, C. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-05 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mitoquinone Inactivates Mitochondrial Chaperone TRAP1 by Blocking the Client Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
4C9T
 
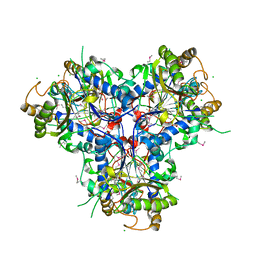 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 2.0 A RESOLUTION, SelenoMet derivative | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RVQ
 
 | | PWI-like domain of Chaetomium thermophilum Brr2 | | Descriptor: | 1,2-ETHANEDIOL, Pre-mRNA splicing helicase-like protein, SULFATE ION | | Authors: | Absmeier, E, Rosenberger, L, Santos, K.F, Becke, C, Wahl, M.C. | | Deposit date: | 2014-11-27 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.135 Å) | | Cite: | A noncanonical PWI domain in the N-terminal helicase-associated region of the spliceosomal Brr2 protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7GF2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MIC-UNK-08cd9c58-1 (Mpro-x11548) | | Descriptor: | 2-(3-chlorophenyl)-N-(1,7-naphthyridin-5-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4CF1
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2s)-6-[[(1r,2s)-2-(5-azanylpentanoylamino)-2,3-dihydro-1h-inden-1-yl]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-13 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CFB
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (R)-(4-CARBOXY-1,3-BENZODIOXOL-5-YL)METHYL-[[2-(CYCLOHEXYLMETHYLCARBAMOYL)PHENYL]METHYL]-METHYL-AZANIUM, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
2CHT
 
 | |
