3IPU
 
 | | X-ray structure of benzisoxazole urea synthetic agonist bound to the LXR-alpha | | Descriptor: | 4-{[methyl(3-{[7-propyl-3-(trifluoromethyl)-1,2-benzisoxazol-6-yl]oxy}propyl)carbamoyl]amino}benzoic acid, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
7JHG
 
 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
1DWI
 
 | |
6AON
 
 | | 1.72 Angstrom Resolution Crystal Structure of 2-Oxoglutarate Dehydrogenase Complex Subunit Dihydrolipoamide Dehydrogenase from Bordetella pertussis in Complex with FAD | | Descriptor: | CALCIUM ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-16 | | Release date: | 2017-08-23 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 2025
|
|
5CKM
 
 | | The CUB1-EGF-CUB2 domains of rat MBL-associated serine protease-2 (MASP-2) bound to Ca2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine peptidase 2 | | Authors: | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | Deposit date: | 2015-07-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
4REB
 
 | | Structural Insights into 5' Flap DNA Unwinding and Incision by the Human FAN1 Dimer | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*GP*CP*GP*AP*GP*CP*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*GP*TP*GP*GP*CP*GP*AP*GP*C)-3'), ... | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
5CCE
 
 | | Joint X-ray/neutron structure of wild type MTAN complexed with SRH and adenine | | Descriptor: | 5'-Methylthioadenosine Nucleosidase, ADENINE, S-ribosylhomocysteine, ... | | Authors: | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | Deposit date: | 2015-07-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4RDU
 
 | |
4RE6
 
 | | Acylaminoacyl peptidase complexed with a chloromethylketone inhibitor | | Descriptor: | Acylamino-acid-releasing enzyme, CHLORIDE ION, N-[(benzyloxy)carbonyl]glycyl-N-[(2S,3R)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]glycinamide | | Authors: | Menyhard, D.K, Orgovan, Z, Szeltner, Z, Szamosi, I, Harmat, V. | | Deposit date: | 2014-09-22 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Catalytically distinct states captured in a crystal lattice: the substrate-bound and scavenger states of acylaminoacyl peptidase and their implications for functionality.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7S6E
 
 | | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Mykhaylyk, V, Orr, C.M, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium
To Be Published
|
|
6UAO
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with the peptide EEYSAM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptide EEYSAM, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
4BYJ
 
 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-(1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
4C71
 
 | | Crystal structure of M. tuberculosis C171Q KasA in complex with TLM18 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(10R)-10-methyloctadecanoyl]oxy}propyl phosphate, (R,E)-3-(4-AZIDOBUTYL)-4-HYDROXY-5-METHYL-5-(2-METHYLBUTA-1,3-DIEN-1-YL)THIOPHEN-2(5H)-ONE, 1,2-ETHANEDIOL, ... | | Authors: | Schiebel, J, Kapilashrami, K, Fekete, A, Bommineni, G.R, Schaefer, C.M, Mueller, M.J, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-09-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the Recognition of Mycolic Acid Precursors by Kasa, a Condensing Enzyme and Drug Target from Mycobacterium Tuberculosis
J.Biol.Chem., 288, 2013
|
|
4QQC
 
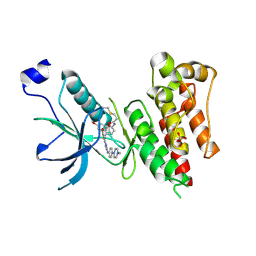 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Domain in Complex with FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6KU0
 
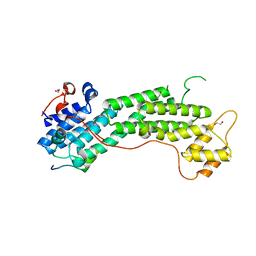 | | Crystal structure of MyoVa-GTD in complex with MICAL1-GTBM | | Descriptor: | 1,2-ETHANEDIOL, Peptide from [F-actin]-monooxygenase MICAL1, Unconventional myosin-Va | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | F-actin disassembly factor MICAL1 binding to Myosin Va mediates cargo unloading during cytokinesis.
Sci Adv, 6, 2020
|
|
6UEG
 
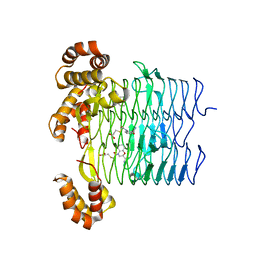 | | Pseudomonas aeruginosa LpxA Complex Structure with Ligand | | Descriptor: | 3-({2-[(2R)-2-carbamoyl-2,3-dihydro-4H-1,4-benzoxazin-4-yl]-2-oxoethyl}sulfanyl)propanoic acid, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CALCIUM ION | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
5CF6
 
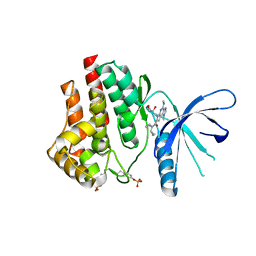 | | CRYSTAL STRUCTURE OF JANUS KINASE 2 IN COMPLEX WITH N,N-DICYCLOPROPYL-10-[(2S)-2,3-DIHYDROXYPROPYL]-3-METHYL-7-(METHYLAMINO)-3,5,8,10-TETRAAZATRICYCLO [7.3.0.02,6]DODECA-1(9),2(6),4,7,11-PENTAENE-11-CARBOXAMIDE | | Descriptor: | N,N-dicyclopropyl-6-[(2S)-2,3-dihydroxypropyl]-1-methyl-4-(methylamino)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Sack, J.S. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Selective Janus Kinase 2 Imidazo[4,5-d]pyrrolo[2,3-b]pyridine Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
6UFA
 
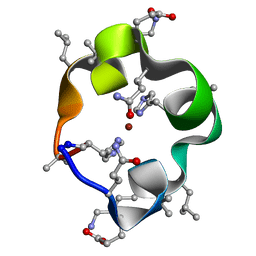 | | S4 symmetric peptide design number 1, Tim zinc-bound form | | Descriptor: | S4-1, Tim, Zinc-bound form, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6AO1
 
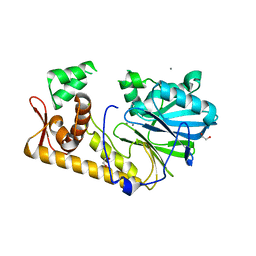 | |
1DWG
 
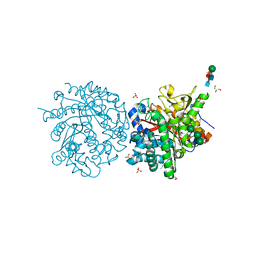 | |
4RED
 
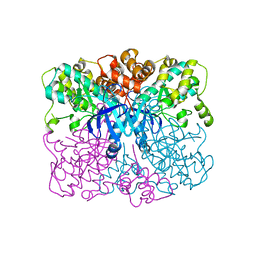 | | Crystal structure of human AMPK alpha1 KD-AID with K43A mutation | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
4REN
 
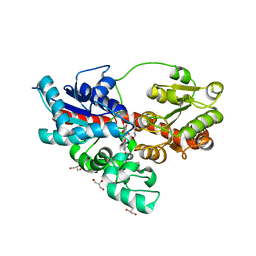 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with petunidin | | Descriptor: | 2-(3,4-dihydroxy-5-methoxyphenyl)-3,5,7-trihydroxychromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
5T5X
 
 | |
2WUG
 
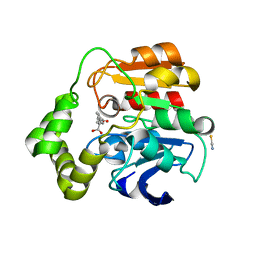 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, ... | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R.L, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a C-C Hydrolase from Mycobacterium Tuberuclosis Involved in Cholesterol Metabolism.
J.Biol.Chem., 285, 2010
|
|
7F5U
 
 | |
